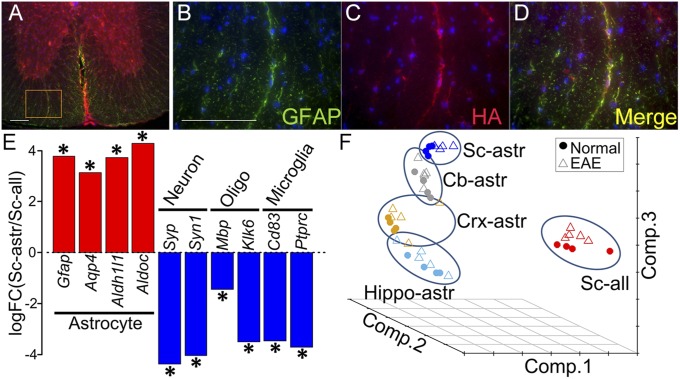
Fig. 1.
Enrichment of astrocyte-specific mRNA using RiboTag technology. (A–D) Double immunolabeling in mGFAP-Cre:RiboTag mice showed colocalization of ribosome associated HA-Tag (red) with the astrocyte-specific marker GFAP (green), with colocalization seen in Merge (yellow). Nuclei were counterstained with DAPI (blue) (40-μm-thick spinal cord sections). B–D are a magnification of Inset area in A. (Scale bars: 100 μm.) (E) Enrichment of astrocyte-specific gene expression and deenrichment of neuronal, oligodendroglial, and microglia-specific gene expression, shown as the log fold change calculated between spinal cord astrocyte RNAs immunoprecipitated by anti-HA antibody versus spinal cord total cell RNAs (including astrocytes) immunoprecipitated with control antibody, anti-RPL22. Asterisks show the significant enrichment or deenrichment (FDR < 0.1). (F) Principal component analysis of astrocyte-enriched RNAs from four different brain regions (Cb, cerebellum; Crx, cerebral cortex; Hippo, hippocampus; and Sc, spinal cord), as well as RNAs from heterogeneous cells from spinal cord (RNA from all cell types). The samples from all cells of spinal cord were markedly separated from astrocyte-enriched spinal cord samples. Within astrocyte-enriched samples, the samples from different brain regions were also separated.