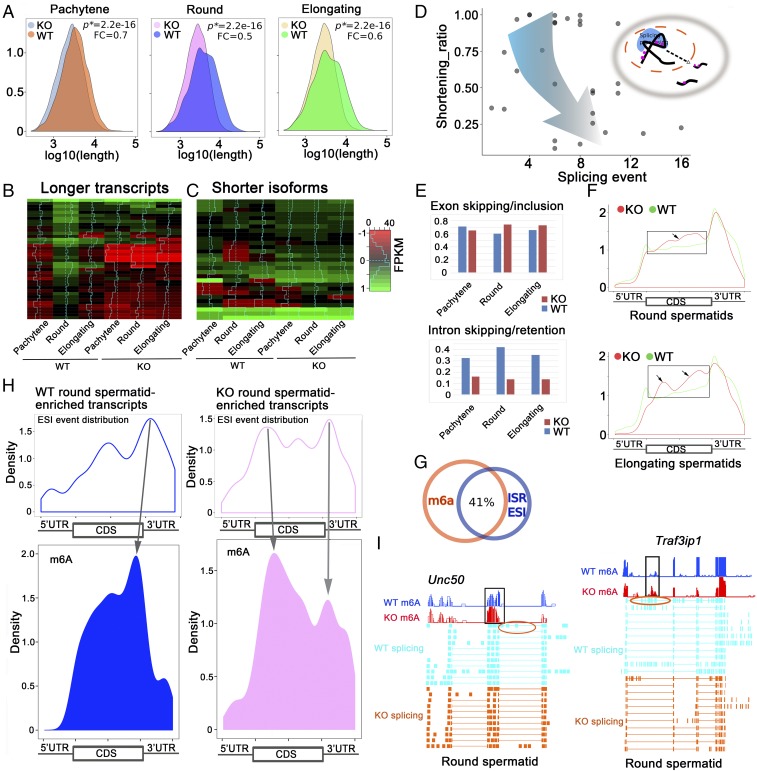
Fig. 3.
Proper m6A erasure is required for the production of longer 3′-UTR mRNAs in pachytene spermatocytes and spermatids. (A) Density plots showing that the total transcript length was significantly decreased in Alkbh5 KO (KO) pachytene spermatocytes and round and elongating spermatids compared with corresponding wild-type (WT) spermatogenic cells. P values of statistical significance (P*) and fold changes (FC) are shown. (B) Heat maps showing the 30 longer 3′-UTR (>3,000 nt) transcripts predominantly expressed in round spermatids in wild-type testes were significantly down-regulated in all three spermatogenic cell types in Alkbh5 KO testes. (C) Heat maps showing that the shorter 3′-UTR isoforms of these 30 longer transcripts were all up-regulated in three spermatogenic cell types in Alkbh5 KO testes compared with wild-type (WT) controls. (D) Dot plot showing the relationship between shortened transcripts and splicing events in the three Alkbh5 KO spermatogenic cell types (pachytene spermatocytes and round and elongating spermatids) analyzed. Shortening ratios (defined as the length of shorter isoform/the length of the longest transcript) were plotted against the number of total splicing events detected based on the RNA-Seq data. The shorter the transcript isoforms become, the more splicing events they tend to have, suggesting that those shorter transcript isoforms were derived from enhanced splicing of those longer transcripts in the three Alkbh5 KO spermatogenic cell types. (E) Histograms showing splicing events, including exon skipping/inclusion and intron skipping/retention, in the three spermatogenic cell types in WT and Alkbh5 KO testes. (F) Comparison of m6A density in transcripts enriched in Alkbh5 KO round (Upper) and elongating (Lower) spermatids with >3 exon skipping/inclusion (ESI) events and those enriched in wild-type (WT) round and elongating spermatids without ESI events. Note that frames indicate elevated m6A levels in the CDS region of the transcripts. (G) Venn diagram showing ∼41% of the m6A sites overlap with the sites with splicing events (±200 nt distance), including exon skipping/inclusion (ESI) and intron skipping/retention (ISR). (H) Density plots showing correlations between splicing and m6A sites in wild-type (WT) and Alkbh5 KO (KO) round spermatid-enriched transcripts. The transcripts enriched in KO round spermatids appear to contain more splicing sites proximal to the m6A sites, compared with those enriched in WT round spermatids, suggesting enhanced splicing events due to m6A accumulation in the KO cells. (I) Examples of two mRNAs (Unc50 and Traf3ip1) up-regulated in Alkbh5 KO round spermatids showing exon skipping events (circled) near the m6A accumulation sites.