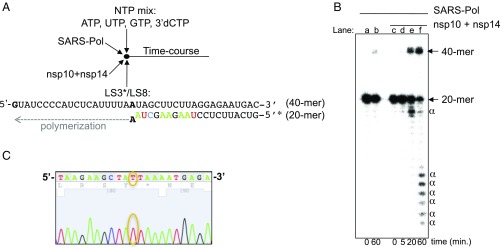
Fig. 5.
In vitro RNA proofreading by the SARS-CoV polymerase complex (SARS-Pol) and exonuclease enzymatic activities. (A) Schematic view of the reaction setup: SARS-Pol (0.5 μM) was incubated with the RNA primer/template LS3*/LS8 (0.5 μM) in the presence of ATP, UTP, GTP, and 3′-dCTP (500 μM each) and, once indicated, with nsp14 (100 nM) and nsp10 (300 nM). Indeed, to prevent dsRNA product degradation by the highly potent nsp10/nsp14-ExoN activity (30), the RNA template bears only a single G at its 5′-end. In the polymerase reaction, only ATP, UTP, GTP and 3′-dCTP are provided as substrates for RNA synthesis. Hence, when the radiolabeled LS3 primer is extended by the polymerase complex, only 3′-dCMP can be incorporated at the 3′-end of the extended primer, preventing nsp10/nsp14-ExoN–mediated degradation of newly synthesized dsRNA (30). Part of the primer is shown with the same color code as the sequencing of the extended primer presented in C. (B) Radiolabeled primer extension by SARS-Pol alone (lane b) or with nsp10 + nsp14 (lanes c–f). RNA products were separated by denaturing gel electrophoresis and analyzed by autoradiography. The positions of the primer (20-mer) and the full-length extension product (40-mer) are indicated. α, dsRNA product degradations by nsp14-ExoN activity. These degradation products may not have biological significance, since replication is uncoupled from excision in the present reconstituted pathway. (C) Part of the Sanger sequencing chromatogram of the extended LS3 from the RT-PCR products formed after incubation of LS3/LS8 with the polymerase complex, nsp10, and nsp14. The surrounding T base corresponds to the corrected base (A to U).