Abstract
Gastric cancer (GC) is the fifth most common malignancy and the third leading cause of cancer-associated mortality worldwide. Therefore, there is a requirement to identify sufficiently sensitive biomarkers for GC. Genome-wide screening of transcriptome dysregulation among cancerous and normal tissues may provide insights into the underlying molecular mechanisms of GC initiation and progression. At present, high-throughput sequencing techniques have begun to innovate biomedical studies. The RNA-seq method has become an advanced approach in medical studies; it is capable of the accurate detection of gene expression levels. The present study used RNA-seq to evaluate the transcriptional changes between tumor and matched normal samples, and these changes were confirmed by differentially expressed genes in larger samples using the results of sequencing. In total, the upregulation of 28 mRNAs and downregulation of 22 mRNAs between cancerous and normal tissue samples were identified. Subsequently, five differentially expressed genes were selected to verify in large samples and cadherin-1 (CDH1) was selected to detect protein expression levels. The results revealed that CDH1, cyclooxygenase-2 and matrix metalloproteinase genes had significantly higher expression levels, whereas the expression levels of dermatopontin and transforming growth factor β receptor 2 were decreased in GC samples. In particular, CDH1 demonstrated a 36-fold higher expression level in cancer tissues. The western blotting results also revealed high CDH1 expression levels in the validation cohorts. Furthermore, these genes are highly enriched in certain gene ontology categories, including the digestive system process, secretion and digestion. The present study provided a preliminary survey of the transcriptome of Chinese patients with GC, which may improve the detection of aberrant gene expression in GC and the understanding of the mechanisms of tumorigenesis.
Keywords: gastric cancer, RNA-seq, differentially expressed gene, gene ontology
Introduction
Gastric cancer (GC) originates in the stomach. It is well established that stomach cancer typically develops gradually over several years in China (1). The initial signs and symptoms include heartburn, upper abdominal pain, nausea and loss of appetite. The advanced case symptoms include weight loss, whiteness of eyes, yellow skin, nausea, difficulty with swallowing and blood in the stool (2). GC may migrate to other organs, including the liver, lungs, bones, lining of the abdomen and lymph nodes (3). Diagnosis is generally determined on the basis of biopsy during endoscopy, and medical imaging technology is used to identify if the disease has progressed to other parts of the body (4). Helicobacter pylori (H. pylori) infection is a leading cause of GC, which accounts for >60% of cases (5), and numerous types of H. pylori, eating pickled vegetables and smoking serve leading roles in the development of GC. Stomach cancer is the fourth most common type of cancer and the second leading cause of cancer-associated mortality worldwide (6). Currently, therapeutic options for the treatment of GC remain inadequate; they include the combination of chemotherapy, radiation therapy, targeted therapy and surgery (7). Therefore, understanding the underlying molecular mechanisms of the carcinogenesis of GC would be useful for designing strategies for the prevention, treatment and control of cancer (8).
A challenge for the application of large-scale functional genomics to cancer research is the identification of the expression profile as a potential source of specific cancer genes that are useful as biomarkers. Previous studies have investigated differentially expressed genes (DEGs) between tumor and normal tissues using high-throughput screening technologies, which to some extent provide numerous possible diagnostic and prognostic biomarkers (9–11). However, GC is a systemic biological disease, and its heterogeneity and the complexity of developing carcinogenesis complicate the diagnosis and determination of its progression (12,13). In the present study, the gap between driver mutations and pathological characteristics of tumor cells are discussed, in addition to facilitating the identification of specific DEGs as potential biomarkers for GC and furthering the understanding of the molecular basis of gene regulation.
The introduction of next-generation sequencing, RNA-seq technology, provides a method for detecting the transcriptome with high precision and at a reasonable cost (14). The present study was designed to illustrate the transcriptome profile in GC tissues and compare it with healthy gastric mucosa using RNA-seq technology. Comparative analyses of gene expression levels were performed to detect DEGs in GC and normal tissues. Thus, the present study generated a large quantity of information on DEGs in Chinese GC tissues vs. normal tissues, which may provide valuable information for evaluation of the underlying molecular mechanism of carcinogenesis, detection of disease markers and the identification of novel targeted anticancer drugs.
Materials and methods
Subject samples
Tissue specimens used in the present study, including tumor and distal normal tissues, were prospectively collected between June 2012 and June 2015 in the Department of Gastroenterology and Surgery, Shanghai Pudong Hospital, Fudan University, Pudong Medical Center (Shanghai, China). The present study included a total of 33 control subjects and 33 patients with GC, who were further divided into the transcriptome profiling groups based on deep sequencing (GC, n=3 patients; controls, n=3 patients) and validation cohorts (GC, n=30; controls, n=30). All patients were Han Chinese. Gastric biopsy samples were obtained from the pyloric antrum of the stomach of control subjects who were referred for upper GI endoscopies due to dyspeptic symptoms, had no previous history of malignancy and no autoimmune or inflammatory disease. GC tissue samples were obtained from surgical specimens immediately following removal from GC patients undergoing primary surgery, with no preoperative irradiation or chemotherapy. Gastric adenocarcinoma in patients with GC was confirmed by histology and classified according to Lauren's criteria into diffuse and intestinal types (15). Demographic and clinical characteristics were collected from all cases and controls using a unified questionnaire. The present study was approved by the Medical Ethics Committee of the Jinshan Hospital of Fudan University. Written informed consent was obtained from all patients prior to enrollment in the present study, conforming to the Declaration of Helsinki.
Tissue sample preparation and RNA extraction
Gastric tissue samples were stored in RNA later (Beijing Solarbio Science and Technology, Co., Ltd., Beijing, China) at 4°C overnight, and then later stored at −80°C. A sample of 30 mg gastric tissue was triturated using liquid nitrogen to homogenize it in sterile conditions. Subsequently, total RNA was extracted from cancerous and normal tissues using TRIzol (Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA, USA), according to the manufacturer's instructions. The quantity and quality (including the ratio of 28S/18S and RNA integrity number) of each RNA sample was assessed using the RNA Nano 6000 assay kit (Agilent Technologies, Inc., Santa Clara, CA, USA) and Bioanalyzer 2100 system (Agilent Technologies, Inc.).
Library preparation and sequencing
A total 10 µg RNA for each sample was used to construct the Illumina sequencing library by the NEBNext mRNA Sample Prep kit 1 (New England Biolabs, Inc., Ipswich, MA, USA). Briefly, total RNA was first selected using oligo-d (T) probes for poly-A mRNAs, then followed by thermal mRNA fragmentation. The fragmented RNA was subjected to cDNA synthesis and further converted into double-stranded cDNA. Upon end repairing, the cDNA product was ligated to Illumina Truseq adaptors and size selected using a 2% agarose gel to generate the average 300 bp cDNA libraries. Polymerase chain reaction (PCR) was subsequently performed using a QIAquick PCR Purification kit (Qiagen GmbH, Hilden, Germany) to determine the relative concentration of the library in order to evaluate the volume to use for sequencing. The RNA-seq library was sequenced on the Illumina Hiseq™ 2500 (Illumina, Inc., San Diego, CA, USA) platform as paired-end (PE) reads to 100 bp using one lane (with a control lane on the same flow cell) at Novogene Bioinformatics Technology Co. Ltd. (Beijing, China). The Digital Gene Expression libraries were generated by Illumina Hiseq™ 2500 with single-end technology in a single run. The average read length of 90 bp was generated as raw data.
Analysis of Illumina transcriptome sequencing results
The Illumina analysis pipeline (CASAVA 1.7) was used to process the raw sequencing data. All raw reads were filtered to remove the adaptor sequence, poly-N reads, low quality reads (50% of the bases had a quality value of ≤5), empty reads (no tags between the adaptors) and reads with only one copy number (possibly due to a sequencing error) and low complexity. Simultaneously, the Q20 (percentage of bases with a Phred value of ≥20) and GC content of the clean data were summarized. Finally, a total of 10 Gbp of cleaned reads was produced. The cleaned sequencing reads were then aligned to the University of California Santa Cruz (UCSC) human reference genome using TopHat software (version 1.0.12; Intel Corporation, Santa Clara, CA, USA), which also incorporates Bowtie software (version 0.11.3; Intel Corporation), to perform the alignment. Burrows-Wheeler Aligner software (version 0.6.1; Intel Corporation) was used to map clean reads to the genome reference (the UCSC human reference genome hg.19), and Bowtie software version 0.11.3 was used for gene referencing. In order to access the transcription abundance for each gene, Cufflinks (version 1.0.3; Intel Corporation) was used to process the aligned reads from alternate samples. The gene transfer format (GTF) file for reference genome annotation used in this analysis was retrieved from the UCSC database. The expression level for each transcript was normalized to the reads per kb of exon model per million mapped reads (FPKM). Cuffdiff (version 1.0.3; Intel Corporation) was used to process the original alignment file produced by TopHat and GTF file for genome annotation to determine the differentially expressed genes. Following evaluation using the Benjamini-Hochberg multiple testing correction, the false discovery rate (FDR) <0.05 was selected as the criteria for significant differences. Gene ontology (GO) was performed and pathway enrichment analysis to investigate the biological significance of the differentially expressed genes. This analysis was performed by the Database for Annotation, Visualization, and Integrated Discovery (DAVID; http://www.biomedsearch.com/nih/DAVID-Database-Annotation-Visualization-Integrated/12734009.html), which is a set of web-based functional annotation tools. The differentially expressed genes and all the expressed genes were submitted as the gene list and background list, respectively. A false discovery rate (FDR) of 1% was used.
The RNA-Seq by Expectation Maximization (RSEM; Illumina, Inc., San Diego, CA, USA) is an RNA-Seq transcript quantification program that currently requires gap-free alignments of RNA-seq reads to Trinity-reconstructed transcripts, including alignments generated by Bowtie software. Given the Trinity-assembled transcripts and the RNA-seq reads generated from a sample, RSEM will directly execute Bowtie to align the reads to the Trinity transcripts and then compute transcript abundance, estimating the number of RNA-seq fragments corresponding to each Trinity transcript, including normalized expression values as FPKM. In addition to estimating the expression levels of individual transcripts, RSEM computes ‘gene-level’ estimates using the Trinity component as a proxy for the gene. To compare the expression levels of different transcripts or genes across samples, a Trinity-included script invokes edge R to perform an additional trimmed mean of M-values (a weighted trimmed mean of the log expression ratios), a scaling normalization that aims to account for differences in total cellular RNA production across all samples. This investigation aimed to identify the different genes between the gastric cancer tissues and normal tissues by sequencing, in order to provide a molecular basis for exploring the pathogenesis of gastric cancer and improve the diagnosis and therapy for gastric cancer.
Reverse transcription-quantitative polymerase chain reaction (RT-qPCR) expression validation
To evaluate the quality of the sequence assembly and expression profile, five differentially expressed genes were selected for amplification, utilizing RT-qPCR. For RT-qPCR, 1 µg total RNA from the transcriptome sample was reverse-transcribed in a 20 µl reaction system, according to the manufacturer's protocol (PrimeScript™ RT Reagent kit; Takara Bio, Inc., Otsu, Japan). The PCR primers were designed based on the sequences of the gene by Primer Premier software (version 5.0, Premier Biosoft International, Palo Alto, CA, USA). Each reaction was carried out in a total volume of 20 µl, with 1 µl cDNA, 10 µl SYBR-Green I Master Mix (LightCycler® 480 SYBR-Green I Master, Roche Diagnostics Ltd., Lewes, UK), 0.5 µl/primer and 9 µl double distilled water. RT-qPCR was performed using the LightCycler® 480 Real-Time PCR system (Roche Diagnostics Ltd.). The RT-qPCR program consisted of 35 cycles of 30 sec at 95°C, 30 sec at 58°C, and 1 min at 72°C, and a final 3 min at 72°C with Premix Taq™ (version 2.0; Takara Biotechnology Co., Ltd.). Each sample was run in triplicate. The data were analyzed with automatic settings for assigning the baseline, and the average Cq and standard deviation (SD) values were calculated. The expression level of mRNA in the tissue was normalized to β-actin. The results were calculated using the ΔΔCT method (16). The primer sequences were as follows: CDH1 forward, AATGCCGCCATCGCTTAC and reverse, TCAGGCACCTGACCCTTGTA; COX-2 forward, GTTCCACCCGCAGTACAGA and reverse, AGGGCTTCAGCATAAAGCGT; MMP-9 forward, TCTATGGTCCTCGCCCTGAA, and reverse, CATCGTCCACCGGACTCAAA; DPT forward, TGTCGCTACAGCAAGAGGTG and reverse, TGAACTTCCACTGGCGATCC; TGFBR2 forward, GCACGTTCAGAAGTCGGATG and reverse, CTGCACCGTTGTTGTCAGTG; β-actin forward, TGCGTGACATTAAGGAGAAG and reverse, GCTCGTAGCTCTTCTCCA.
Western blot analysis
Gastric tissue from the control and case samples was homogenized in 300 ml lysis buffer [50 mM Hepes; pH 7.5; 150 mM NaCl; 10 mM EDTA; 10 mM glycerophosphate; 100 mM sodium fluoride; 1% Triton X-100; 1 mM phenylmethane sulfenylfluoride and protease inhibitor (PI) cocktail]. Following centrifugation of the homogenate (20,000 × g, 15 min) the supernatants were used for western blotting. A total of 50 µg protein extracts from samples were suspended in Laemmli buffer (100 mM Hepes; pH 6.8; 10% β-mercaptoethanol; 20% SDS), incubated at 100°C for 5 min to denature the proteins, and loaded onto a 10% gel to undergo SDS-PAGE. Following separation, proteins were electrically transferred onto a nitrocellulose membrane. The membrane was incubated with blocking solution (1X TBS; 0.05% Tween-20; 5% non-fat milk) at room temperature for 1 h and incubated overnight with primary antibodies raised against CDH1 (cat. no. 14472; 1:1,000; Cell Signaling Technology, Inc., Danvers, MA, USA). Following incubation with the corresponding horseradish peroxidase-conjugated rat anti-mouse IgG secondary antibodies for 1 h at room temperature (cat. no. sc-130300; 1:5,000; Santa Cruz Biotechnology, Inc., Dallas, TX, USA), proteins were visualized using an Enhanced Chemiluminescence Plus Immunoblotting Detection system (GE Healthcare Life Sciences, Chalfont, UK). The intensity of the immunoreactive bands was quantified using a blot analysis system (Bio-Rad Laboratories, Inc., Hercules, CA, USA), and β-actin was used as a loading control. Commercial markers (SeeBlue pre-stained standard; Invitrogen; Thermo Fisher Scientific, Inc.) were used as molecular weight standards. Each experiment was repeated three times.
Statistical analysis
The FPKM data was analyzed using the Pearson's correlation coefficient, the paired t-test and the Benjamini Hochberg correction for false discovery rate, such that the differential expression was considered to be significant with a difference of P<0.01. The data was normalized using rank invariant normalization and analyzed using the High-Throughput qPCR package in R (version 3.3.3; https://www.bioconductor.org/) and Bioconductor (version 3.5; https://www.bioconductor.org/). The validation and plasma RT-qPCR expression data was analyzed using the nonparametric Mann-Whitney U-test. P<0.05 was considered to indicate a statistically significant difference for the Benjamini Hochberg adjustment.
Results
Transcriptome sequencing
The cancer and normal tissue samples were subjected to massively parallel primer extension (PE) cDNA sequencing. A total of 178.2 and 175.4 million raw reads were obtained of 100 bp length in the normal and cancer tissues from Illumina sequencing, respectively. Subsequently, the raw reads were filtered by removing low quality reads and reads containing N and adaptor sequences. The remaining reads were termed ‘clean reads’ and used for downstream bioinformatic analysis. Burrows-Wheeler Aligner software (version 0.6.1; Intel Corporation) was used to map clean reads to the genome reference (the UCSC human reference genome hg.19), and Bowtie software was used for gene referencing. The unique match reads for subjected samples were 154.5 and 151.4 million raw reads. The average coverage of sequencing depth was ~600 times the human transcriptome (30 Mbp and ~1% of the hg.19, based on the total length of the uniquely annotated exon region in the Ensembl database; http://www.ensembl.org/Homo_sapiens/Location/View?db=core;g=ENSG00000231978;r=1:21768269-21768575;t=ENST00000434488). The results are presented in Table I.
Table I.
RNA sequencing results of mRNA.
| Sample name | Total reads (%) | Clean reads (%) | Genome map rate (%) | Gene map rate (%) |
|---|---|---|---|---|
| Control 1 | 59,318,462 (100) | 51,352,541 (86.6) | 79.24 | 73.17 |
| Control 2 | 60,891,232 (100) | 52.065,542 (85.48) | 78.41 | 72.41 |
| Control 3 | 58,010,790 (100) | 51.167,261 (88.21) | 77.53 | 71.62 |
| Case 1 | 57,390,348 (100) | 49.765,172 (85.91) | 76.33 | 73.61 |
| Case 2 | 58,540,531 (100) | 51.152,986 (87.25) | 78.12 | 74.51 |
| Case 3 | 59,643,118 (100) | 50.351,271 (85.12) | 77.29 | 72.65 |
Differentially expressed genes
Subsequently, the present study detected the gene expression levels and identified the differentially expressed genes between case and control samples using the software package, RSEM. RSEM computes maximum likelihood abundance estimates using the Expectation-Maximization (EM) algorithm for its statistical model, including the modeling of PE and variable-length reads, fragment length distributions and quality scores, to determine which transcripts are isoforms of the same gene. The FPKM method was used to determine the gene expression levels. In total, 14,318 and 14,694 expressed genes were evaluated by postulating that the FPKM value was >1 among any samples of each group. The analysis contained the majority of the annotated human genes. Following this, the correlation of the gene expression between two samples was evaluated. The results revealed that gene expression levels among samples were highly correlated (Pearson's correlation coefficient r=0.92), suggesting that the experiments were reliable and the samples chosen were reasonable (Table II).
Table II.
Correlation values between samples.
| Sample | Control 1 | Control 2 | Control 3 | Case 1 | Case 2 | Case 3 |
|---|---|---|---|---|---|---|
| Control 1 | 1.000 | 0.997 | 0.982 | 0.991 | 0.984 | 0.989 |
| Control 2 | 0.994 | 1.000 | 0.978 | 0.000 | 0.985 | 0.993 |
| Control 3 | 0.995 | 0.995 | 1.000 | 0.985 | 0.982 | 0.994 |
| Case 1 | 0.986 | 0.995 | 0.981 | 1.000 | 0.987 | 0.987 |
| Case 2 | 0.994 | 0.997 | 0.972 | 0.971 | 1.000 | 0.973 |
| Case 3 | 0.997 | 0.993 | 0.993 | 0.975 | 0.989 | 1.000 |
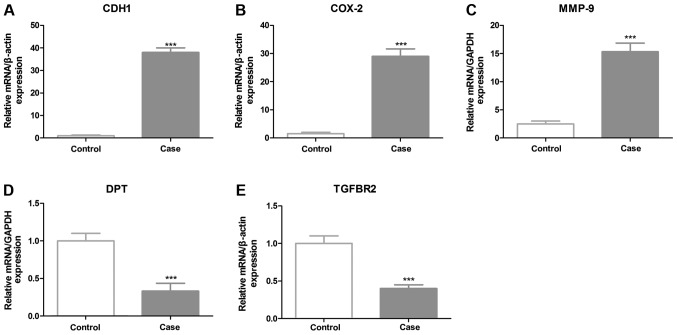
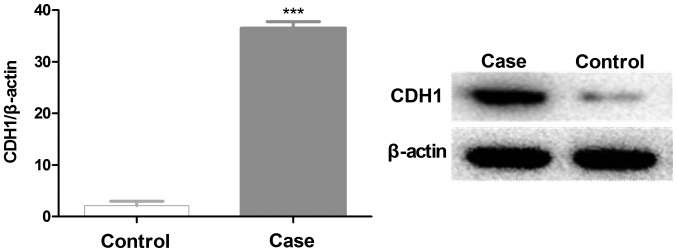
In order to refine the analysis, the selection criteria was strengthened with a threshold of FDR ≤0.05 and fold-change ≥3 applied. The stringent criteria generated a list of 28 instances of mRNA upregulation and 22 instances of downregulation between cancer and normal samples, which are presented in Fig. 1. The genes ranked with the five highest expression levels between normal and cancer samples are as follows: Cadherin-1 gene (CDH1), cyclooxygenase 2 (COX-2), matrix metalloproteinase-9 (MMP-9), dermatopontin (DPT) and transforming growth factor β receptor II (TGFBR2). Further evaluation confirmed high-throughput sequencing in validation cohorts, including samples that participated in deep sequencing. A total of five genes expressed in normal and cancer samples were selected to be verified by RT-qPCR. The results revealed that CDH1, COX-2 and MMP-9 had significantly increased expression levels, whereas the expression levels of DPT and TGFBR2 were decreased in GC samples, compared with the control (Fig. 2). Notably, CDH1 demonstrated a 36-fold higher expression level in cancer tissues. In previous studies CDH1 has been revealed to have significantly altered expression levels in GC tissues. The most significantly downregulated gene in cancer tissue samples is DPT, which encodes the dermatopontin protein. Furthermore, western blot analysis results also demonstrated that CDH1 was highly expressed in validation cohorts (Fig. 3).
Figure 1.
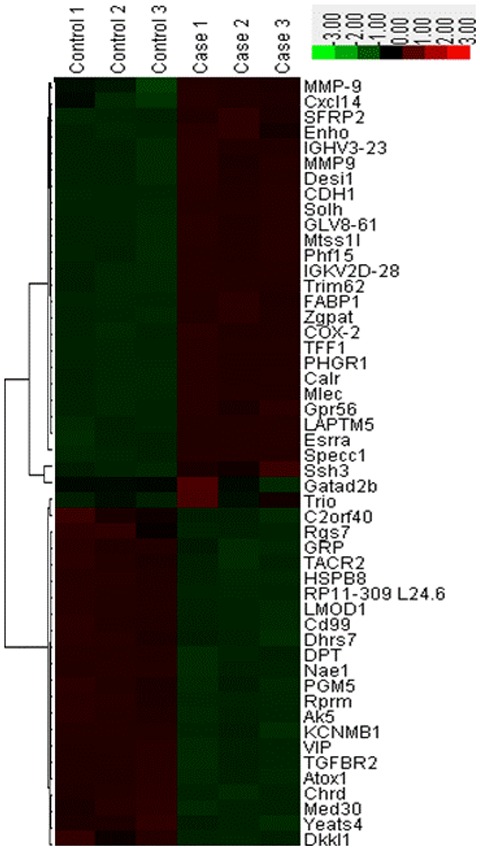
Heat map and cluster analysis of the 50 differentially expressed mRNAs in control and gastric cancer tissue samples. Red represents high relative expression levels; Green represents low relative expression levels.
Figure 2.
(A-E) Validation of differentially expressed mRNA in larger cohorts (GC, n=30; control, n=30). The expression levels of five mRNAs in case and control subjects were determined by RT-qPCR. β-actin was used as a normalization control. All reactions were based on three independent repetitions. RT-qPCR, reverse transcription-quantitative polymerase chain reaction; CDH1, cadherin-1; COX-2, cyclooxygenase 2; MMP-9, matrix metalloproteinase-9; DPT, dermatopontin; TGFBR2, transforming growth factor β receptor II. ***P<0.001 compared with the control group.
Figure 3.
Quantification by western blot analysis of the CDH1 protein expression levels in control and case samples (GC, n=30; control, n=30). CDH1, cadherin-1. ***P<0.001 compared with the control group.
Functional enrichment analysis of DEGs
To better understand the biological function of DEGs, GO enrichment analysis was performed. GO is an international standard gene functional classification system that offers a dynamic-updated controlled vocabulary, in addition to a strictly defined concept to comprehensively describe properties of genes and their products in organisms. GO has three ontologies: Molecular function, cellular component and biological process. The basic unit of GO is a GO-term. Every GO-term belongs to a type of ontology. GO enrichment analysis provides all GO terms that significantly enriched in a list of DEGs, comparing to a genome background, and filter the DEGs that correspond to specific biological functions. This method firstly maps all DEGs to GO terms in the database (http://www.geneontology.org/), calculating gene numbers for each term; subsequently a hypergeometric test was performed to identify significantly enriched GO terms in the input list of DEGs based on GO: TermFinder. In the present study, only biological process and molecular function categories were considered. The functional enrichment work was performed using an online tool, DAVID. Using a threshold of FDR <0.05, it was revealed that all differentially expressed genes were categorized into 12 functional categories (Table III). For example, the over-represented GO categories include digestive system process, regulation of body fluid levels, secretion and digestion.
Table III.
Enriched GO categories of DEGs.
| Category | GO ID | GO term | Cluster frequency | Genome frequency of use | Corrected P-value |
|---|---|---|---|---|---|
| BP | GO:0022600 | Digestive system process | 4/50 genes, 8% | 19/15,332 genes, 0.1%a | 0.0012 |
| GO:0050878 | Regulation of body fluid | 6/50 genes, 12% | 835/15,332 genes, 5.4%a | 0.0018 | |
| GO:0046903 | Secretion | 8/50 genes, 16% | 143/15,332 genes, 0.9%a | 0.0024 | |
| GO:0007586 | Digestion | 5/50 genes, 10% | 253/15,332 genes, 1.7%a | 0.0027 | |
| GO:0007155 | Cell adhesion | 13/50 genes, 26% | 373/15,332 genes, 2.4%a | 0.0040 | |
| GO:0022610 | Biological adhesion | 14/50 genes, 28% | 2,954/15,332 genes, 19.3%a | 0.0041 | |
| GO:0007267 | Cell-cell signaling | 14/50 genes, 28% | 3,787/15,332 genes, 24.7%a | 0.0031 | |
| GO:0007967 | System process | 13/50 genes, 26% | 3,787/15,332 genes, 24.7%a | 0.0029 | |
| MF | GO:0005184 | Neuropeptide hormone | 6/50 genes, 12% | 24/15,332 genes, 0.2%a | 0.0147 |
| GO:0005509 | Calcium ion binding | 20/50 genes, 40% | 4,489/15,451 genes, 29.1%a | 0.0152 | |
| GO:0005179 | Cytokine activity | 10/50 genes, 20% | 3,537/15,451 genes, 22.9%a | 0.0703 | |
| GO:0003823 | Symporter activity | 12/50 genes, 24% | 2,111/15,451 genes, 27%a | 0.0431 |
P<0.05 vs. the control group. GO, gene ontology; DEG, differentially expressed gene; ID, identity; BP, biological process; MF, molecular function.
Discussion
The present study comprehensively investigated the transcriptome in GC and normal tissue samples, and further verified DEG expression levels in large samples. By applying the whole transcriptome sequencing technology (RNA-seq), it was estimated that the expression levels of DEGs were associated with GC tissues. Additionally, the present study also provided novel ideas for the identification of underlying molecular mechanisms of GC and its role in the diagnosis and treatment of GC.
An Illumina Hiseq 2500 platform with a 90-bp sequencing read length was used in the present study. A total of >350 million raw reads were obtained; this number of raw reads has been used in previous studies to achieve sufficient sequencing coverage for transcriptome profiling. A total of 98% sequencing reads that align with the hg19 met the quality standards of the RNA-seq technique. Therefore, the mRNA-seq data provided a sufficient profile of expressed genes in the subject genome. Furthermore, the present study further verified RNA-seq data in large samples by RT-qPCR and western blotting. Results generated from RT-qPCR and western blotting were consistent with the high-throughput sequencing results, which indicates that high-throughput sequencing may be a novel approach for the precision treatment of cancer.
The present study verified numerous DEGs and isoforms in gene expression levels in GC tissues. A number of the genes identified are known to be involved in various types of cancer. Based on the results of the present study, 12 GO categories were confirmed to over-represent among the DEGs, including in the regulation of body fluid levels, secretion and digestive system process categories (Table III). Consistent with previous studies, transcriptome analysis in the present study identified DEGs with biological functions that were associated with cell adhesion. The CDH1 gene demonstrated the most significant difference between normal and cancer tissue samples, with a 36-fold higher expression in GC tissues. Similarly, the expression level of CDH1 protein was also significantly higher in GC tissues (Fig. 3). The CDH1 gene is associated with the cadherin superfamily (17–19): Sequence mutations occurring in this gene are always correlated with GC. Loss of function may contribute to the progression of GC by increasing proliferation, invasion and/or metastasis (20–24), which is associated with cancer progression, metastasis and decreased level of cellular adhesion in the tissue, ultimately increasing cellular motility (25–29).
COX-2 has a length of 8,3 kb and is positioned on chromosome 1 (1q25.2–25.3), and consists of 10 exons and 9 introns, encoding 603 or 604 amino acids; However, it is not expressed under normal physiological conditions (30–34). A number of previous studies have revealed that the COX-2 gene was one of the early growth response genes and may be widely vessels inside and outside activator (including interleukin 15-serotonin transforming growth factor), resulting in tumorigenesis (35–39). Sun et al (40) revealed that in superficial gastritis (100%), atrophic gastritis (35.7%), intestinal metaplasia (37.8%), stomach dysplasia (41.7%) and GC (69.5%), of COX-2 positive the expression was gradually increasing trend. COX-2 is considered to be an initial process in gastric carcinogenesis.
MMP-9 is a class of enzymes associated with the zinc-MMP family complex and the degradation of the extracellular matrix. The MMP-9 gene encodes a signal peptide in humans, a propeptide; it has a catalytic domain, followed by three repeats of a fibronectin type II domain and finally a C-terminal hemopexin-like domain (41). Specifically, MMP-9 is associated with the pathogenesis of cancer, due to its key role in extracellular matrix remodeling and angiogenesis; its elevated expression level has been previously demonstrated in metastatic mammary cancer cell lines. Gelatinase B serves a role in tumor progression and angiogenesis, stromal remodeling and eventually metastasis (42). It may be challenging to utilize gelatinase B inhibition in cancer therapy strategies due to its physiological function. However, the role of gelatinase B in tumor metastasis diagnosis has previously been studied: Complexes of gelatinase B/tissue inhibitors of metalloproteinases were revealed to have increased expression levels in gastrointestinal cancer and gynecologic malignancies. MMPs, including MMP-9, may participate in the progression of numerous human malignancies, as the degradation of collagen IV in the basement membrane and extracellular matrix assists tumor progression, including invasion, metastasis, growth and angiogenesis (43). A number of studies detected that MMP-9 expression in GC tissues was higher, compared with in normal gastric mucosa, and serosal invasion, and MMP-9 persons were significantly higher compared with those without serosal invasion, as determined by flow cytometry (44–46). The differentiated group of lymph node metastasis was revealed to have a higher frequency than those deprived of lymph node metastasis (P≤0.05). Additionally, Shan et al (47) demonstrated suppressed expression levels of MMP-9 in a poorly segregated GC cell line, BGC-823, by pGenesil carrie, which subsequently set the foundation for further in vivo experiments and gene therapy (47,48).
Dermatopontin is a protein, which is encoded by the DPT gene and is present in numerous tissues types. Briefly, it is an extracellular matrix protein with possible functions in cell-matrix interactions and matrix assembly, and a number of its tyrosine residues are sulfated. It has also been suggested that dermatopontin may modify the behavior of TGF β by interacting with decorin (49). The TGFBR2 gene encodes a member of the serine/threonine protein kinase family receptor. These encoded proteins have a protein kinase domain, which forms a heterodimeric complex with other protein receptors and binds TGF-β. These receptor/ligand complexes phosphorylate proteins, prior to entering the nucleus and regulating the transcription of genes associated with the cell proliferation process. TGFBR2 gene mutations have previously been associated with Marfan syndrome, Loeys-Deitz aortic aneurysm syndrome, Osler-Weber-Rendu syndrome and the development of numerous types of tumors (50,51). Furthermore, spliced transcript variants encoding for a number of isoforms have also been characterized (52,53).
In summary, based on RNA-seq technology, sequencing reads were generated to profile the GC transcriptome. The results provided a wealth of information on DEGs in case-control tissue samples, which may benefit further studies and lead to improved methods for GC detection and therapy.
Acknowledgements
The present study was supported by the Training Program Foundation for Talent of the Jinshan Hospital of Fudan University (Shanghai, China; grant no. JHFU201326148).
References
- 1.Chen W, Zheng R, Baade PD, Zhang S, Zeng H, Bray F, Jemal A, Yu XQ, He J. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66:115–132. doi: 10.3322/caac.21338. [DOI] [PubMed] [Google Scholar]
- 2.Patel SA. The inferior vena cava (IVC) syndrome as the initial manifestation of newly diagnosed gastric adenocarcinoma: A case report. J Med Case Rep. 2015;9:204. doi: 10.1186/s13256-015-0696-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Du T, Zhang B, Zhang S, Jiang X, Zheng P, Li J, Yan M, Zhu Z, Liu B. Decreased expression of long non-coding RNA WT1-AS promotes cell proliferation and invasion in gastric cancer. Biochim Biophys Acta. 2016;1862:12–19. doi: 10.1016/j.bbadis.2015.10.001. [DOI] [PubMed] [Google Scholar]
- 4.Yao K, Nagahama T, Matsui T, Iwashita A. Detection and characterization of early gastric cancer for curative endoscopic submucosal dissection. Dig Endosc. 2013;25(Suppl 1):S44–S54. doi: 10.1111/den.12004. [DOI] [PubMed] [Google Scholar]
- 5.Amorim I, Smet A, Alves O, Teixeira S, Saraiva AL, Taulescu M, Reis C, Haesebrouck F, Gärtner F. Presence and significance of Helicobacter spp. in the gastric mucosa of Portuguese dogs. Gut Pathog. 2015;7:12. doi: 10.1186/s13099-015-0057-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ferlay J, Soerjomataram I, Dikshit R, Eser S, Mathers C, Rebelo M, Parkin DM, Forman D, Bray F. Cancer incidence and mortality worldwide: Sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136:E359–E386. doi: 10.1002/ijc.29210. [DOI] [PubMed] [Google Scholar]
- 7.Delaunoit T. Latest developments and emerging treatment options in the management of stomach cancer. Cancer Manag Res. 2011;3:257–266. doi: 10.2147/CMAR.S12713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yegin EG, Kani T, Banzragch M, Kalayci C, Bicakci E, Duman DG. Survival in patients with hypoechoic muscularis propria lesions suggestive of gastrointestinal stromal tumors in gastric wall. Acta Gastroenterol Belg. 2015;78:12–17. [PubMed] [Google Scholar]
- 9.Printz C. High-throughput sequencing detects signs of cancer recurrence. Cancer. 2012;118:4097. doi: 10.1002/cncr.27780. [DOI] [PubMed] [Google Scholar]
- 10.Conley A, Minciacchi VR, Lee DH, Knudsen BS, Karlan BY, Citrigno L, Viglietto G, Tewari M, Freeman MR, Demichelis F, Di Vizio D. High-throughput sequencing of two populations of extracellular vesicles provides an mRNA signature that can be detected in the circulation of breast cancer patients. RNA Biol. 2017;14:305–316. doi: 10.1080/15476286.2016.1259061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dallol A, Buhmeida A, Al-Ahwal MS, Al-Maghrabi J, Bajouh O, Al-Khayyat S, Alam R, Abusanad A, Turki R, Elaimi A, et al. Clinical significance of frequent somatic mutations detected by high-throughput targeted sequencing in archived colorectal cancer samples. J Transl Med. 2016;14:118. doi: 10.1186/s12967-016-0878-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shaheen MF, Barrette P. Successful endoscopic management of gastric perforation caused by ingesting a sharp chicken bone. Int J Surg Case Rep. 2015;9:12–14. doi: 10.1016/j.ijscr.2015.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bonfrate L, Grattagliano I, Palasciano G, Portincasa P. Dynamic carbon 13 breath tests for the study of liver function and gastric emptying. Gastroenterol Rep (Oxf) 2015;3:12–21. doi: 10.1093/gastro/gou068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Luo YH, Liang L, He RQ, Wen DY, Deng GF, Yang H, He Y, Ma W, Cai XY, Chen JQ, Chen G. RNA-sequencing investigation identifies an effective risk score generated by three novel lncRNAs for the survival of papillary thyroid cancer patients. Oncotarget. 2017;8:74139–74158. doi: 10.18632/oncotarget.18274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tichá V, Jansa P, Tichý M. Retrospective study of gastric cancer on the basis of Lauren's classification criteria. Acta Univ Palacki Olomuc Fac Med. 1988;120:193–198. [PubMed] [Google Scholar]
- 16.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 17.Chen QH, Deng W, Li XW, Liu XF, Wang JM, Wang LF, Xiao N, He Q, Wang YP, Fan YM. Novel CDH1 germline mutations identified in Chinese gastric cancer patients. World J Gastroenterol. 2013;19:909–916. doi: 10.3748/wjg.v19.i6.909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Garziera M, Canzonieri V, Cannizzaro R, Geremia S, Caggiari L, De Zorzi M, Maiero S, Orzes E, Perin T, Zanussi S, et al. Identification and characterization of CDH1 germline variants in sporadic gastric cancer patients and in individuals at risk of gastric cancer. PloS One. 2013;8:e77035. doi: 10.1371/journal.pone.0077035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Garziera M, De Re V, Geremia S, Seruca R, Figueiredo J, Melo S, Simões-Correia J, Caggiari L, De Zorzi M, Canzonieri V, et al. A novel CDH1 germline missense mutation in a sporadic gastric cancer patient in north-east of Italy. Clin Exp Med. 2013;13:149–157. doi: 10.1007/s10238-012-0184-7. [DOI] [PubMed] [Google Scholar]
- 20.Hansford S, Kaurah P, Li-Chang H, Woo M, Senz J, Pinheiro H, Schrader KA, Schaeffer DF, Shumansky K, Zogopoulos G, et al. Hereditary diffuse gastric cancer syndrome: CDH1 mutations and beyond. JAMA Oncol. 2015;1:23–32. doi: 10.1001/jamaoncol.2014.168. [DOI] [PubMed] [Google Scholar]
- 21.de Campos EC, Ribeiro S, Higashi R, Manfredini R, Kfouri D, Cavalcanti TC. Hereditary diffuse gastric cancer: laparoscopic surgical approach associated to rare mutattion of CDH1 gene. Arq Bras Cir Dig. 2015;28:149–151. doi: 10.1590/s0102-67202015000200017. (In English, Portuguese) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.van der Post RS, Vogelaar IP, Manders P, van der Kolk LE, Cats A, van Hest LP, Sijmons R, Aalfs CM, Ausems MG, Gómez García EB, et al. Accuracy of hereditary diffuse gastric cancer testing criteria and outcomes in patients with a germline mutation in CDH1. Gastroenterology. 2015;149:897–906.e19. doi: 10.1053/j.gastro.2015.06.003. [DOI] [PubMed] [Google Scholar]
- 23.Benusiglio PR, Colas C, Rouleau E, Uhrhammer N, Romero P, Remenieras A, Moretta J, Wang Q, De Pauw A, Buecher B, et al. Hereditary diffuse gastric cancer syndrome: Improved performances of the 2015 testing criteria for the identification of probands with a CDH1 germline mutation. J Med Genet. 2015;52:563–565. doi: 10.1136/jmedgenet-2015-103153. [DOI] [PubMed] [Google Scholar]
- 24.Lajus TB, Sales RM. CDH1 germ-line missense mutation identified by multigene sequencing in a family with no history of diffuse gastric cancer. Gene. 2015;568:215–219. doi: 10.1016/j.gene.2015.05.035. [DOI] [PubMed] [Google Scholar]
- 25.Lynch HT, Lynch JF. Hereditary diffuse gastric cancer: lifesaving total gastrectomy for CDH1 mutation carriers. J Med Genet. 2010;47:433–435. doi: 10.1136/jmg.2010.078170. [DOI] [PubMed] [Google Scholar]
- 26.Matsukuma KE, Mullins FM, Dietz L, Zehnder JL, Ford JM, Chun NM, Schrijver I. Hereditary diffuse gastric cancer due to a previously undescribed CDH1 splice site mutation. Hum Pathol. 2010;41:1200–1203. doi: 10.1016/j.humpath.2010.01.022. [DOI] [PubMed] [Google Scholar]
- 27.Pinheiro H, Bordeira-Carriço R, Seixas S, Carvalho J, Senz J, Oliveira P, Inácio P, Gusmão L, Rocha J, Huntsman D, et al. Allele-specific CDH1 downregulation and hereditary diffuse gastric cancer. Hum Mol Genet. 2010;19:943–952. doi: 10.1093/hmg/ddp537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen B, Zhou Y, Yang P, Liu L, Qin XP, Wu XT. CDH1-160C>A gene polymorphism is an ethnicity-dependent risk factor for gastric cancer. Cytokine. 2011;55:266–273. doi: 10.1016/j.cyto.2011.04.008. [DOI] [PubMed] [Google Scholar]
- 29.Chen Y, Kingham K, Ford JM, Rosing J, Van Dam J, Jeffrey RB, Longacre TA, Chun N, Kurian A, Norton JA. A prospective study of total gastrectomy for CDH1-positive hereditary diffuse gastric cancer. Ann Surg Oncol. 2011;18:2594–2598. doi: 10.1245/s10434-011-1648-9. [DOI] [PubMed] [Google Scholar]
- 30.Zhang P, Luo HS, Li M, Tan SY. Artesunate inhibits the growth and induces apoptosis of human gastric cancer cells by downregulating COX-2. Onco Targets Ther. 2015;8:845–854. doi: 10.2147/OTT.S81041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yu F, Li K, Chen S, Liu Y, Li Y. Pseudolaric acid B circumvents multidrug resistance phenotype in human gastric cancer SGC7901/ADR cells by downregulating Cox-2 and P-gp expression. Cell Biochem Biophys. 2015;71:119–126. doi: 10.1007/s12013-014-0170-7. [DOI] [PubMed] [Google Scholar]
- 32.Bai YN, Zhang P, Li L, Wang SL, Yao NL, Zhang RS, Liu Z, Yan D, Zhu YL, Ma JZ, et al. Effect of jianpi tongluo jiedu recipe on expression levels of COX-2, NF-kappaBp65, and Bcl-2 in gastric mucosa of patients with precancerous lesions of gastric cancer. Zhongguo Zhong Xi Yi Jie He Za Zhi. 2015;35:167–173. (In Chinese) [PubMed] [Google Scholar]
- 33.Aziz F, Yang X, Wang X, Yan Q. Anti-LeY antibody enhances therapeutic efficacy of celecoxib against gastric cancer by downregulation of MAPKs/COX-2 signaling pathway: Correlation with clinical study. J Cancer Res Clin Oncol. 2015;141:1221–1235. doi: 10.1007/s00432-014-1892-z. [DOI] [PubMed] [Google Scholar]
- 34.Wang Z, Chen JQ, Liu JL. COX-2 inhibitors and gastric cancer. Gastroenterol Res Pract. 2014;2014:132320. doi: 10.1155/2014/132320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schildberg C, Abbas M, Merkel S, Agaimy A, Dimmler A, Schlabrakowski A, Croner R, Leupolt J, Hohenberger W, Allgayer H. COX-2, TFF1, and Src define better prognosis in young patients with gastric cancer. J Surg Oncol. 2013;108:409–413. doi: 10.1002/jso.23416. [DOI] [PubMed] [Google Scholar]
- 36.Chen Z, Liu M, Liu X, Huang S, Li L, Song B, Li H, Ren Q, Hu Z, Zhou Y, Qiao L. COX-2 regulates E-cadherin expression through the NF-kappaB/Snail signaling pathway in gastric cancer. Int J Mol Med. 2013;32:93–100. doi: 10.3892/ijmm.2013.1376. [DOI] [PubMed] [Google Scholar]
- 37.Tseng YC, Tsai YH, Tseng MJ, Hsu KW, Yang MC, Huang KH, Li AF, Chi CW, Hsieh RH, Ku HH, Yeh TS. Notch2-induced COX-2 expression enhancing gastric cancer progression. Mol Carcinog. 2012;51:939–951. doi: 10.1002/mc.20865. [DOI] [PubMed] [Google Scholar]
- 38.Targosz A, Brzozowski T, Pierzchalski P, Szczyrk U, Ptak-Belowska A, Konturek SJ, Pawlik W. Helicobacter pylori promotes apoptosis, activates cyclooxygenase (COX)-2 and inhibits heat shock protein HSP70 in gastric cancer epithelial cells. Inflam Res. 2012;61:955–966. doi: 10.1007/s00011-012-0487-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shin WG, Kim HJ, Cho SJ, Kim HS, Kim KH, Jang MK, Lee JH, Kim HY. The COX-2-1195AA genotype is associated with diffuse-type gastric cancer in Korea. Gut Liver. 2012;6:321–327. doi: 10.5009/gnl.2012.6.3.321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sun WH, Yu Q, Shen H, Ou XL, Cao DZ, Yu T, Qian C, Zhu F, Sun YL, Fu XL, Su H. Roles of Helicobacter pylori infection and cyclooxygenase-2 expression in gastric carcinogenesis. World J Gastroenterol. 2004;10:2809–2813. doi: 10.3748/wjg.v10.i19.2809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zucker S, Lysik RM, DiMassimo BI, Zarrabi HM, Moll UM, Grimson R, Tickle SP, Docherty AJ. Plasma assay of gelatinase B: tissue inhibitor of metalloproteinase complexes in cancer. Cancer. 1995;76:700–708. doi: 10.1002/1097-0142(19950815)76:4<700::AID-CNCR2820760426>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 42.Groblewska M, Siewko M, Mroczko B, Szmitkowski M. The role of matrix metalloproteinases (MMPs) and their inhibitors (TIMPs) in the development of esophageal cancer. Folia Histochem Cytobiol. 2012;50:12–19. doi: 10.5603/FHC.2012.0002. [DOI] [PubMed] [Google Scholar]
- 43.Li M, Yang G, Xie B, Babu K, Huang C. Changes in matrix metalloproteinase-9 levels during progression of atrial fibrillation. J Int Med Res. 2014;42:224–230. doi: 10.1177/0300060513488514. [DOI] [PubMed] [Google Scholar]
- 44.Avci N, Ture M, Deligonul A, Cubukcu E, Olmez OF, Sahinturk S, Topak A, Kurt E, Evrensel T, Şahin AB, Yakut T. Association and prognostic significance of the functional −1562C/T polymorphism in the promoter region of MMP-9 in Turkish patients with gastric cancer. Pathol Oncol Res. 2015;21:1243–1247. doi: 10.1007/s12253-015-9950-7. [DOI] [PubMed] [Google Scholar]
- 45.Xia Y, Lian S, Khoi PN, Yoon HJ, Joo YE, Chay KO, Kim KK, Do Jung Y. Chrysin inhibits tumor promoter-induced MMP-9 expression by blocking AP-1 via suppression of ERK and JNK pathways in gastric cancer cells. PloS One. 2015;10:e0124007. doi: 10.1371/journal.pone.0124007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Akter H, Park M, Kwon OS, Song EJ, Park WS, Kang MJ. Activation of matrix metalloproteinase-9 (MMP-9) by neurotensin promotes cell invasion and migration through ERK pathway in gastric cancer. Tumour Biol. 2015;36:6053–6062. doi: 10.1007/s13277-015-3282-9. [DOI] [PubMed] [Google Scholar]
- 47.Shan YQ, Ying RC, Zhou CH, Zhu AK, Ye J, Zhu W, Ju TF, Jin HC. MMP-9 is increased in the pathogenesis of gastric cancer by the mediation of HER2. Cancer Gene Ther. 2015;22:101–107. doi: 10.1038/cgt.2014.61. [DOI] [PubMed] [Google Scholar]
- 48.Shimura T, Dagher A, Sachdev M, Ebi M, Yamada T, Yamada T, Joh T, Moses MA. Urinary ADAM12 and MMP-9/NGAL complex detect the presence of gastric cancer. Cancer Prev Res (Phila) 2015;8:240–248. doi: 10.1158/1940-6207.CAPR-14-0229. [DOI] [PubMed] [Google Scholar]
- 49.Yamatoji M, Kasamatsu A, Kouzu Y, Koike H, Sakamoto Y, Ogawara K, Shiiba M, Tanzawa H, Uzawa K. Dermatopontin: A potential predictor for metastasis of human oral cancer. Int J Cancer. 2012;130:2903–2911. doi: 10.1002/ijc.26328. [DOI] [PubMed] [Google Scholar]
- 50.Docea AO, Mitruţ P, Grigore D, Pirici D, Călina DC, Gofiţă E. Immunohistochemical expression of TGF beta (TGF-beta), TGF beta receptor 1 (TGFBR1), and Ki67 in intestinal variant of gastric adenocarcinomas. Rom J Morphol Embryol. 2012;53(Suppl 3):S683–S692. [PubMed] [Google Scholar]
- 51.Shinto O, Yashiro M, Kawajiri H, Shimizu K, Shimizu T, Miwa A, Hirakawa K. Combination effect of a TGF-beta receptor kinase inhibitor with 5-FU analog S1 on lymph node metastasis of scirrhous gastric cancer in mice. Cancer Sci. 2010;101:1846–1852. doi: 10.1111/j.1349-7006.2010.01606.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Park D, II, Son HJ, Song SY, Choe WH, Lim YJ, Park SJ, Kim JJ, Kim YH, Rhee PL, Paik SW, et al. Role of TGF-beta 1 and TGF-beta type II receptor in gastric cancer. The Korean J Intern Med. 2002;17:160–166. doi: 10.3904/kjim.2002.17.3.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Park SH, Kim YS, Park BK, Hougaard S, Kim SJ. Sequence-specific enhancer binding protein is responsible for the differential expression of ERT/ESX/ELF-3/ESE-1/jen gene in human gastric cancer cell lines: Implication for the loss of TGF-beta type II receptor expression. Oncogene. 2001;20:1235–1245. doi: 10.1038/sj.onc.1204227. [DOI] [PubMed] [Google Scholar]