Abstract
The presence of androgen receptor variant 7 (AR-V7) variants becomes a significant hallmark of castration resistant prostate cancer (CRPC) relapsed from hormonal therapy and is associated with poor survival of CRPC patients because of lacking a ligand-binding domain. Currently, it still lacks an effective agent to target AR-V7 or AR-Vs in general. Here, we showed a novel class of agents (thailanstatins, TSTs, spliceostatin A analogs) can significantly suppress the expression of AR-V7 mRNA and protein but in a less extent on the full-length AR expression. Mechanistically, TST-D is able to inhibit AR-V7 gene splicing by interfering the interaction between U2AF65 and SAP155 and preventing them from binding to polypyrimidine tract located between the branch point and the 3' splice site. In vivo, TST-D exhibits a potent tumor inhibitory effect on human CRPC xenografts leading to cell apoptosis. The machinery associated with AR gene splicing in CRPC is a potential target for drugs. Based on their potency in the suppression of AR-V7 responsible for the growth/survival of CRPC, TSTs representing a new class of anti-AR-V agents warrant further development into clinical application.
Keywords: gene splicing, AR variants, thailanstatin, CRPC
1. INTRODUCTION
Prostate cancer (PCa) has been the second common cause of cancer deaths in men only behind lung cancer in the past two decades [1]. Androgen receptor (AR) plays a central role in PCa development [2]; even in castration resistant PCa (CRPC) cells, AR or AR-mediated signaling pathways remain active [3]. Multiple mechanisms leads to hyperactive AR signaling, including AR mutation or amplification, altered expression of AR co-activators and co-repressors, AR activation through crosstalk with other signaling pathways, or presence of AR splice variants (AR-Vs [4; 5; 6; 7; 8]). Therefore, there remains a critical and unmet need for novel therapeutic agents that can target AR signaling in CRPC.
For patients with metastatic PCa, androgen deprivation therapy (ADT) is the standard treatment. The majority of advanced PCa patients respond to initial ADT temporarily, but they inevitably progress from an androgen-dependent stage to CRPC [9]. Currently, ADT is to either reduce androgen production by surgical or chemical castration or inhibit AR activity by blocking androgen binding to the C-terminal ligand-binding domain (LBD) of AR (i.e., anti-androgen) in which docking of AR antagonists into LBD results in the passive AR inhibition via competition with agonists. Although there are new available ADT agents such as cytochrome P450 17 (CYP17) inhibitor Abiraterone [10] and the androgen-receptor antagonist Enzalutamide (formerly known as MDV-3100) [11], ADT is not curative and only prolongs the relapse of CRPC. Surprisingly, recent studies have identified several constitutively active AR-Vs lacking the LBD [12]. In this case, even complete androgen ablation would have no effect on AR-V activity. In addition, AR-Vs also play a role in mechanism of resistance to chemotherapies such as Taxanes [13]. AR-Vs are refractory to traditional treatment[11; 14], which highlights the need for a new class of AR-inhibitory agents for successful management of tumors expressing truncated ARs or AR-Vs. For example, the active AR inhibition may either prevent co-activator(s) binding, or induce co-repressor(s) recruitment to the N-terminal transactive domain, or suppress the expression of AR or AR-Vs.
Currently, it is hypothesized that the formation of AR-Vs can occur through two different mechanisms: genomic rearrangement and alternative splicing. Genomic rearrangement of AR gene has been reported in PCa cell lines and clinical tissues [15; 16], but it’s still controversial because RNA-Seq studies did not detect it in the SU2C series [17]. Alternative splicing of AR precursor mRNA (pre-mRNA) is common mechanism for generating a variety of AR-V proteins expressed in PCa cell lines and specimens[18]. It is known that more than 90% of human genes undergo programmed alternative splicing to generate protein variants far greater than the ~30,000 encoding genes [19]. While this process inherently provides for transcriptome diversity, aberrant alternative splicing has been implicated in numerous disease conditions such as cancer and neurodegeneration[20; 21]. In malignancies, alternative splicing of the pre-mRNAs of oncogenes is frequent mechanism used by the cell to generate protein variants that have oncogenic activity [21]. Thus, targeting aberrant pre-mRNA alternative splicing represents a new opportunity and strategy to eradicate recurrent CRPC expressing AR-Vs. In this study, we explored novel spliceosome inhibitors as potential therapeutic agents using CRPC model in vivo and identified their mechanism of action in vitro.
2. MATERIALS AND METHODS
Cell culture, antibodies, and reagents
PNT1A, LAPC-4, 22RV1, PZ-HPV7, RWPE-1, and HEK293 were purchased from American Type Culture Collection (ATCC, Manassas, VA) and were maintained in medium according to ATCC instruction. C4-2 was originally generated by our lab. In addition, LN95 cell was provided by Dr. Jun Luo (Johns Hopkins Medical School) and maintained in Phenol red-free RPMI 1640 (Life Technologies, Carlsbad, CA) with 10% Charcoal-stripped Fetal bovine serum (CS-FBS). C4-2 and PNTA1 were maintained in RPMI 1640 (Life Technologies) supplemented with 10% FBS. LAPC-4 was cultured in Iscove’s-Dulbecco’s Modified Eagle’s Medium (I-DMEM; Life Technologies) with 10% FBS. VCaP was generously provided by Dr. Ralf Kittler (UT Southwestern Medical Center) and maintained in Dulbecco’s Modified Eagle’s Medium (DMEM; Life Technologies) containing 10% FBS. Cell lines were passaged in our laboratory for fewer than 40 times after resuscitation. Cell lines were authenticated using AmpFLSTR®Identifier® PCR Amplification kit (Life Technologies) and confirmed as mycoplasma-free using MycoAlert® kit (Lonza Walkersville, Inc. Walkersville, MD). All the antibodies used in this study were purchased from several commercial sources listed in Supplemental Table S1.
Preparation of thailanstatins from Burkholderia thailandensis MSMB43
Thailanstatin A (TST-A; Fig. S1) was purified to at least 95% homogeneity from the fermentation broth of Burkholderia thailandensis MSMB43 as previously described[22]. Thailanstatin D (TST-D), an analogue of TST-A without a hydroxyl group on the heavily substituted tetrahydrofuran ring (Fig. S1), was newly discovered and purified similarly as TST-A from the fermentation broth of an engineered bacterial strain[23]. Diaion HP-20 resin (Sigma-Aldrich, St. Louis, MO) was a key reagent used for the purification of TSTs. The final compound preparations were verified by mass spectrometry (MS) and by nuclear magnetic resonance (NMR) analysis.
Crystal violet assay for in vitro cell growth
For determining the effect of TSTs on cell growth, 4,000 cells were seeded per well in 96-well plates. After 24 hours, cells were treated with different concentrations of TST-A or TST-D. At the indicated time, cells were washed in PBS once, fixed in 100 µl/well 1% glutaraldehyde (Sigma-Aldrich) for 15 minutes, after the removal, then stained with 100 µl/well 0.5% crystal violet (Sigma-Aldrich) for 15 minutes. After washing out excess dye, 100 µl/well Sorenson’s solution was added to elute dye from stained cells and subjected to densitometry measurement at OD560 nm. The relative cell number was determined using control =1.
Plasmids and cell transfection
Human AR-V7 cDNA was obtained from Dr. Jun Luo (Johns Hopkins Medical School) and AR-V7 minigene was provided by Dr. Xuesen Dong (Vancouver Prostate Centre, University of British Columbia). Flag-tagged U2AF65 and hnRNP I plasmids were obtained from Dr. James Manley (Columbia University) and Dr. Allain Fre´de´ ric (Institute for Molecular Biology and Biophysics Eidgenȍssiche Technische Hochshule, Switzerland). All the plasmid transfections were performed with Lipofectamine 3000 transfection kit (Life Technologies) according to the manufacturer's instructions.
Western blot and immunoprecipitation (IP)
Cells were harvested and lysed using ice-cold lysis buffer [150 mM NaCl, 1% Triton X-100, 0.5% sodium deoxycholate, 0.1% SDS, 50 mM Tri (pH 8.0), protease inhibitor cocktail (Roche, Indianapolis, IN)] for 45 min or subjected to nuclear and cytoplasmic extraction (NE-PER; Pierce Biotechnology, Rockford, IL). Lysates were centrifuged at 13,000 rpm for 10 minutes at 4°C. Equivalent amounts of protein were separated on 4–12% gradient NuPAGE Bis-Tris Plus Gels (Life Technologies) and transferred (Trans-Blot® SD Semi-Dry Electrophoretic Transfer Cell, Bio-Rad, Hercules, CA) to nitrocellulose membranes. Membranes were blocked in TBS-Tween 20 containing 3% non-fat dry milk (w/v) for 1 hour and then incubated with primary antibodies overnight at 4°C. Appropriate secondary antibodies conjugated with horseradish peroxidase and enhanced chemiluminescence (Pierce Biotechnology) with Supersignal® West Dura Extended Duration Substrate kit (Thermo Fisher Scientific, Waltham, MA) were used to detected target proteins. For IP, the immunocomplexes were firstly precipitated with Dynabeads Protein G (Life Technologies) and then subjected to western blot analysis. Bands were addressed by densitometric scans and showed after normalizing to Actin.
Quantitative real-time PCR (qRT-PCR)
Total RNA was extracted with Maxwell 16 LEV simplyRNA tissue kit (Promega, Madison, WI) and cDNA was made from 1 µg RNA using iScript Advanced cDNA Synthesis kit (Bio-Rad, Hercules, CA). qRT-PCR analysis was set up with SYBR Green qPCR Supermix kit (Life Technologies) and carried out in iCycler thermal cycler (Bio-Rad). The relative level of mRNA expression of each gene was determined by normalizing with 18S rRNA. All experiments were repeated at least twice in duplicates. The primers used in this experiment were listed in Supplemental Table S2.
RNA pull-down assay
RNA oligo (CUACAGUCAACAAUGUCUCUCUUUCAUACUAGAAAAAUUC) was synthesized (Life Technologies) and biotinylated using Pierce RNA 3´-end Desthiobiotinylation Kit (Thermo Fisher Scientific). Biotinylated RNA oligo (50 pmol) was incubated with streptavidin magnetic beads (Thermo Fisher Scientific) at room temperature for 30 minutes and immobilized RNA oligo was incubated with 80 µg extracted nuclear protein in a binding buffer containing 50% glycerol and 50 mM NaCl at 4°C for 2 hours. The beads were washed three times and proteins were eluted then subjected to MS or western blot analysis.
Mouse xenografts, histology examination, and immunohistochemistry (IHC) staining
22RV1 mixed with Matrigel (Thermo Fisher Scientific) was injected subcutaneously. Once tumor becomes palpable, TST-D (300 µg/kg/day) or DMSO (control) was delivered using ALZET osmotic pumps (DURECT Corporation, Cupertino, CA). Tumor volume was determined prior and post-injection and each tumor was excised and weighted then subjected to histological examination and target validation using western blot. All animal protocols were approved by the Institutional Animal Care and Use Committee in UT Southwestern Medical Center.
Hematoxylin and eosin (H&E) and IHC staining for cleaved Caspase-3 were carried out in formalin-fixed, paraffin-embedded tissue sections with 5–6 µm sections. For IHC, sections were subjected to heat-induced antigens retrieval (Tris/EDTA, pH 9.0) for 30 minutes at 97°C °Cthen to Dako envision detection using DAKO autostainer Link48 (Dako, Carpinteria, CA).
Statistical analysis
All error bars in graphical data represent mean ± SD. Student’s two-tailed t-test was used for the determination of statistical relevance between groups, and P<0.05 was considered statistically significant. All statistical analyses were performed with GraphPad Prism software.
3. RESULTS
The effect of TSTs on prostate epithelial cells
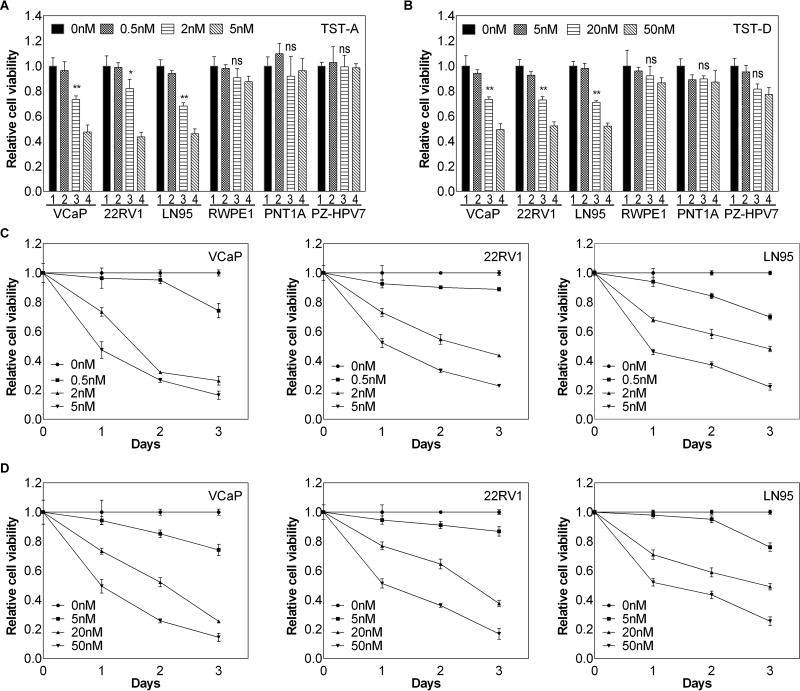
FR901464 (FR), a bacterial natural product from a Pseudomonas species, and spliceostatin A (SSA), a methylated derivative of FR, are prototypic pre-mRNA splicing inhibitors and exhibited potent anti-proliferative activities [24]. However, the inherent chemical instability of FR and SSA has hindered their development. Recently, we identified several natural FR analogues, namely thailanstatins (TST-A, TST-B, TST-C and TST-D) [22] and unpublished data, from an exotic bacterial species Burkholderia thailandensis MSMB43. TST-A and TST-D retain an intact epoxide group critical for their potent in vitro pre-mRNA splicing inhibitory activity (IC50 in single µM range) and anti-proliferative activity against human cancer cell lines (IC50 in single nM range). Although TST-A appeared to be 10 times more potent than TST-D, TST-A is less stable than TST-D. We therefore compared the effect of both TST-A and TST-D on different prostate cell lines. As shown in Fig. 1A and B, TST-A and TST-D (Fig. S1A) exhibited a significant growth inhibition in all 3 CRPC cell lines (i.e., VCaP, 22RV1 and LN95) expressing both AR and AR-V7; IC50 is around 5 nM or 50 nM for TST-A or TST-D respectively. In contrast, either agent exhibited little effect on immortalized normal prostate epithelial cells (such as RWPE-1, PNTA1 and PZ-HPV-7) (Fig. 1A and 1B) or other PCa cells (such as C4-2 and LAPC-4) expressing only AR but not AR-V7 (Fig. S1B). In addition, both agents inhibited the cell growth of these 3 CRPC cell lines in both dose- and time-dependent manners (Fig. 1C and D).
Figure 1. The effect of TSTs on prostate cell lines.
Different prostate cell lines were treated with TST-A (A) or TST-D (B) at different concentrations and relative cell number was determined by crystal violet assay 24 hours after treatment. The kinetics of TST-A (C) or TST-D (D) on three CRPC cell lines (i.e., 22RV1, VCaP and LN95)-expressing AR-V7 was determined subsequently. * P<0.05 ** P<0.01
The effect of TST-A and TST-D on the expression of full-length AR (AR-FL) and AR-V7
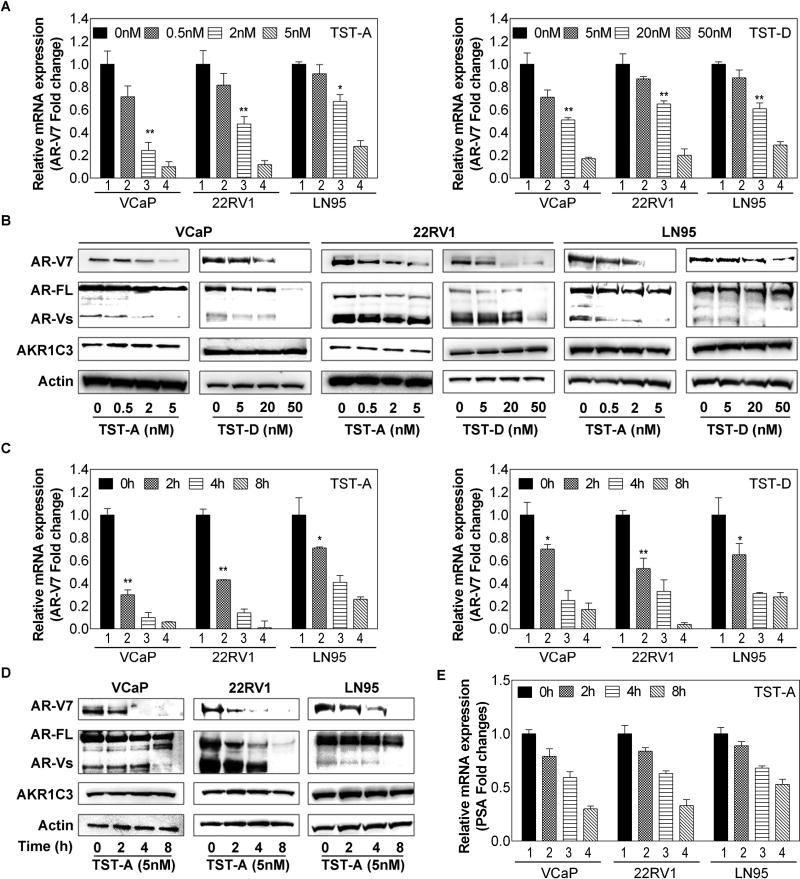
Knowing TST as a potent splicing inhibitor, we therefore determined the steady-state levels of AR-V7 mRNA in VCaP, 22RV1 and LN95 cells treated with different concentrations of TST-A or TST-D. As shown in Fig. 2A and S2A, the mRNA levels of both AR-V7 and AR-FL were suppressed by TST-A or TST-D in a dose-dependent manner. Similar to mRNA results, the protein levels of AR-V7 and other AR-Vs were significantly decreased with the incremental concentrations of TST-A or TST-D (Fig. 2B). Also, immunostaining data indicated that nuclear AR-V7 disappeared in treated cells (Fig. S3). In addition, time course data indicated that the inhibitory effect of TSTs on AR-V7 (Fig. 2C) or AR-FL (Fig. S2B) mRNA expression could be detected as early as 2 hours after treatment. Accordingly, AR-V7 protein was reduced (Fig. 2D) but less for AR-FL protein (Fig. S2B) that may be due to the longer half-life of AR-FL protein [25]. Nevertheless, the effect of TSTs on AR expression appeared to be gene specific after examining several other genes (Fig. S4) with alternative splicing variants that did not show any consistent inhibition.
Figure 2. The effect of TSTs on AR gene expression in CRPC cell lines.
Cells were treated with TST-A at 0, 0.5, 2, 5 nM (A) or TST-D at 0, 5, 20, 50 nM (B) for 4 hours, total cellular RNA and cell lysates were prepared and subjected to western blot and qRT-PCR analyses. The relative AR-V7 mRNA expression from each treatment was normalized with 18S rRNA then calculated using control (=1). Cells were treated with 5 nM TST-A or 50 nM TST-D, cell pellets were collected 0, 2, 4, 8 hours after treatment. Total cellular RNA and cell lysates were prepared and subjected to qRT-PCR analyses (C) or western blot (D).
Meanwhile, PSA expression as a reporter of AR activity was also detected by PCR. As shown in Fig 2E, PSA reduction could be detected 2 hours after treatment. We further examined the effect of TST on gene without alternative splicing variants such as AKR1C3 (a steroidogenesis enzyme in androgen synthesis) and both PCR and western results (Fig. 2A and S2C) showed that the expression of AKR1C3 was not impacted by TSTs. It appeared that the inhibitory effect of TSTs was mainly on AR gene expression, which could be due to the actively expression of AR gene in CRPC cell.
AR-V7 as a major target of TST-D-elicited cytotoxicity
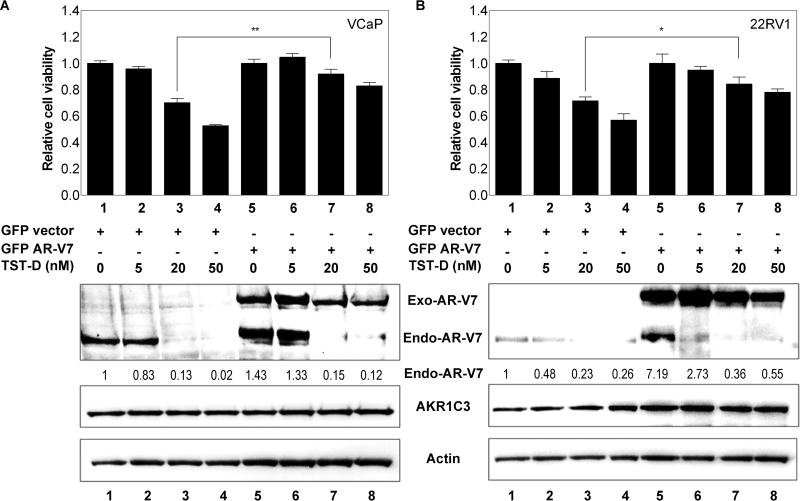
In order to delineate the role of AR-V7 or AR-FL in TST-elicited growth inhibition of CRPC, both 22RV1 and VCAP cells were transiently transfected with exogenous AR-V7 or AR-FL cDNA vector prior to drug treatment. As shown in Fig 3A and B, elevated AR-V7 expression in 22RV1 and VCAP could significantly reduce cell death caused by TST-D (by comparing lane 3 and lane 7) at 20nM. However, exogenous AR-FL cDNA expression failed to rescue cells from TST-D cytotoxicity as AR-V7 cDNA (Fig. S5); these results also indicated that CRPC cells become addictive to the presence of AR-V7 for its growth advantage. Furthermore, western blot analyses further confirmed that exogenous AR-V7 or AR-FL expression was not affected by TST-D while only endogenous AR-V7 or AR-FL was significantly decreased by TST-D (Fig. 3 and S5), suggesting the effect of TSTs on AR gene splicing.
Figure 3. The effect of exogenous AR-V7 cDNA expression on TST cytotoxicities.
22RV1 (A) and VCaP (B) cells were transfected with GFP-vec or GFP-AR-V7 cDNA for 24 hours, cells were subjected to different concentrations of TST-D. For determining cell growth, cells were harvested 24 hours after treatment (upper panel). For analyzing protein expression, cells were collected 4 hours after treatment (lower panel). * P<0.05, ** P<0.01.
The mechanism of TST action on AR gene splicing
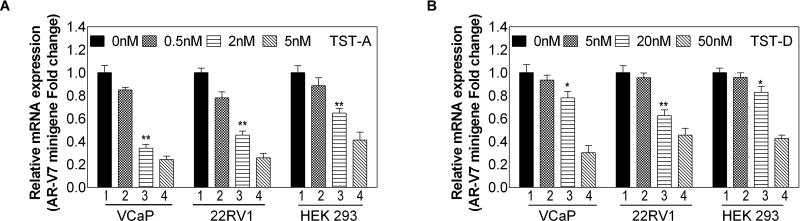
To unveil the mechanism of TSTs on AR gene splicing, AR minigene [26] was employed as a splicing reporter vector in 22RV1 and VCaP cells to determine the effect of TSTs, AR-V7 or AR-FL can be detected using different primers. In addition, HEK293 cell without endogenous AR expression was used as well. As shown in Fig. 4 and Fig. S6, the expression of cDNA products that mimic AR-V7 and AR-FL were decreased in a dose-dependent manner with treatment of TST-A and TST-D, which is similar to the effect of TSTs on inhibiting the endogenous AR gene expression.
Figure 4. The mechanism of TST action on AR gene splicing.
22RV1, VCaP and HEK293 cells were transiently transfected with mock vector or ARV7 minigene plasmid. 24 hours after transfection, cells were treated with TST-A at 0, 0.5, 2, 5 nM (A) or TST-D at 0, 5, 20, 50 nM (B) for 4 h respectively. Relative mRNA levels of AR-V7 mRNA levels to 18S rRNA were determined by real-time qPCR.
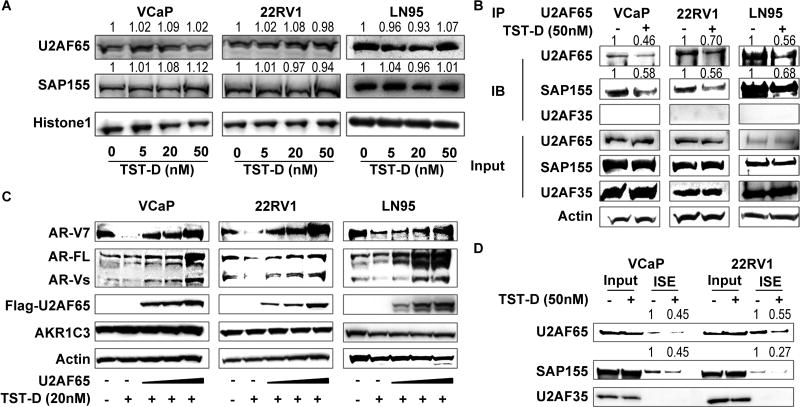
Since TSTs can effectively inhibit the expression of AR-V7 minigene that contains intronic splicing enhancers (ISE) critical for the formation of AR-V7 mRNA, ISE binding protein, U2AF65, has been demonstrated to play an essential role in AR-V7 pre-mRNA splicing[26] by interacting with other splicing factors such as SF3B complex during pre-mRNA splicing. We therefore used ISE in a RNA pull-down assay and conducted MS analysis to identify the potential splicing factors that are necessary in AR-V7 splicing. SAP155 (also called SF3B1), U2AF65, and U2AF35 appeared to be potential candidates (Supplemental Table S3). We first surveyed the effect of TST-D on the expression of both U2AF65 and SAP155 in VCAP, 22RV1 and LN95, and found that TST-D did not alter the steady-state levels of expression of either protein (Fig. 5A), however, TST-D can block the interaction between U2AF65 and SAP155. Furthermore, by transfecting U2AF65 cDNA expression vector into VCaP, 22RV1, and LN95 cells, we observed that exogenous U2AF65 expression could recover splicing procedure and upregulate AR or AR-V7 expression in TST-D treated cells in a dose-dependent manner. Under the same experimental conditions, we noticed that the expression of neither AKR1C3 nor actin was affected, suggesting that gene splicing of both genes is not highly active in these cells (Fig. 5C). Meanwhile, we carried out RNA pull-down assay using synthetic RNA oligonucleotides representing AR-V7 ISE to determine whether TST-D could decrease the binding of splicing factor to ISE clearly indicated that less binding of U2AF65/SAP155 complex to ISE of AR-V7 in the presence of TST-D (Fig. 5D)
Figure 5. The mechanism of TST action on AR gene splicing.
(A) VCaP, 22RV1 and LN95 were treated with TST-D at 0, 5, 20, 50 nM for 24 hours; the expression of both U2AF65 and SAP155 proteins was analyzed by western blot. Histone 1 was used as the loading control. (B) 22RV1, VCaP and LN95 cells were pre-treated with 50 nM TST-D for 4 hours; total cell lysate were immunoprecipitated with U2AF65 antibodies plus protein G beads. Bound proteins were eluted and analyzed by western blot. (C) 22RV1, VCaP and LN95 cells were transiently transfected with mock vector or Flag-U2AF65 plasmid. Forty-eight hours after transfection, cells were treated with 20 nM TST-D for 4 hours. Both AR-V7 and AR-FL were analyzed by western blot. (D) 22RV1 and VCaP cell lysates were incubated with RNA oligos containing ISE. Oligo-associated factors were eluted and subjected to western blot for determining the interaction of splicing factor(s) with AR-V7 ISE.
Recent data [27, 28] indicated that U2AF35 is a critical factor for exon skipping. However, it didn’t appear to be involved in AR-V7 splicing (Fig. 5B and S7). In addition, RNA pull-down (Fig 5D) and MS data (Supplemental Table S3) indicated that U2AF35 did not directly associate with U2AF65/SAP155 complex nor interact with ISE implying that U2AF35 are not involved in AR-V7 gene splicing. Collectively, these data conclude that the mechanism of TST-D is to target the assembly of U2AF65/SAP155 complex that is critical for the splicing of AR-V7 pre-mRNA.
The evaluation of in vivo efficacy of TST-D in CRPC therapy
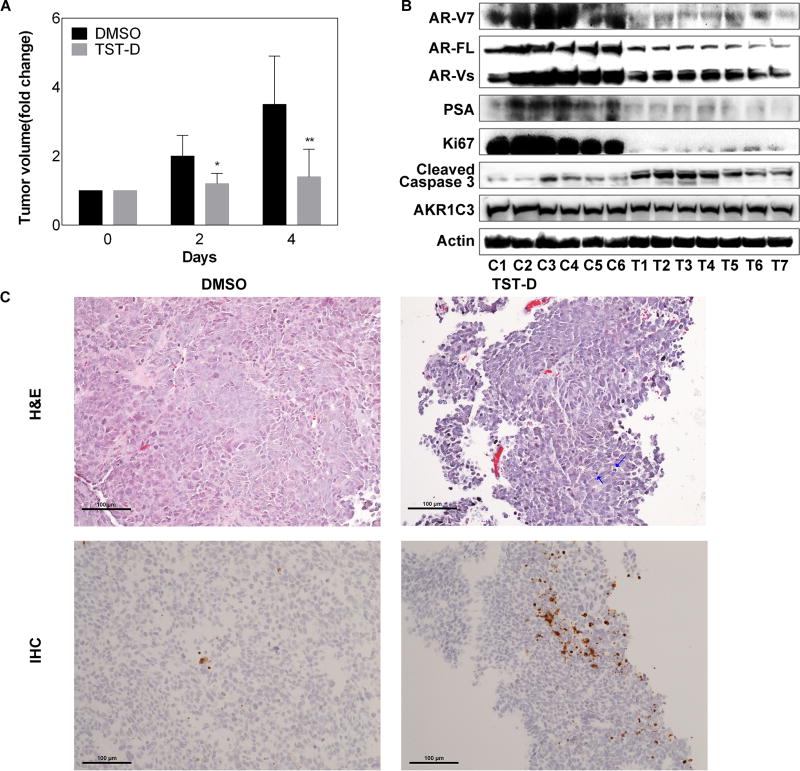
To examine the in vivo effect of TSTs, we employed a xenograft model using Enzalutamide-resistant 22RV1 [25] and preliminarily tested TST-D in this study. As shown in Fig. 6A and S8A, TST-D significantly inhibited the tumor growth. TST-D treatment was able to suppress the protein expression of AR-V7 (or other AR-Vs) as well as prostate specific antigen (PSA) in a dose-dependent manner (Fig. 6B and S8B), reflecting overall AR activities. Similar to the in vitro data (Fig. 2B), AKR1C3 remained unchanged in tumor cells during the course of treatment. The apoptotic marker such as cleaved Caspase-3 was significantly elevated but proliferation maker such as Ki67 protein was significantly decreased in the treated tumors (Fig. 6B). Histologic examination also unveiled that the treated tumors exhibited significant drug-elicited cell apoptosis compared with control (Fig. 6C). Consistent with Western blot data, the IHC result indicated that the apoptotic marker such as cleaved Caspase-3 was significantly elevated in the treated tumors (Fig. 6C). Taken together, these results suggested that TST-D could be a promising agent to target CRPC tumor expressing AR-Vs.
Figure 6. The in vivo effect of TST-D on CRPC xenograft.
(A) Mice bearing 22RV1 xenografts were treated with DMSO or 300 µg/kg/day TST-D; tumor volumes were measured every other day and tumor growth rate from each tumor was calculated based on pre-treatment. (B) The protein expression of AR-V7, AR-FL, PSA, Ki67, cleaved Caspase-3 and AKR1C3 from tumors collected 4 days after treatment was determined by western blot. C: control; T: treatment. (C) H&E staining was employed to examine drug-elicited cell death and confirmed by IHC staining using cleaved Caspase-3 antibodies (lower panel). Apoptic cells are marked with arrow. * P<0.05 ** P<0.01
4. DISCUSSION
In the past few years, several new agents have been approved for metastatic CRPC patients[10; 11], but those new agents, such as the CYP17 inhibitor (i.e., Abiraterone), second-generation AR antagonist (i.e., Enzalutamide), and an androgen receptor N-terminal domain antagonist EPI-001 [29] still target androgen synthesis and AR because AR remains active even in CRPC. Despite of overwhelming response to these new regimens, the recurrent cancer eventually relapses resulted in the mortality of this disease. Therefore, the mechanism(s) leading to drug resistance become the focal point of combating CRPC. Noticeably, the appearance of several AR-Vs in CRPC cells after ADT becomes one of the suspicious causes of the failure of ADT. Truncated AR-Vs without LBD such as AR-V7 are functional active to promote androgen-independent growth for CRPC cells; most significantly, all anti-AR agents have no effect on these proteins. As expected, the presence of AR-V7 in circulating tumor cells from CRPC patients is highly correlated with patient survival [30].
Currently, several studies have attempted to develop some new drugs to target AR-Vs. For example, AR degradation compound ASC-J9 [31], BRD4 inhibitor JQ1[32] and CYP17 inhibitor galeterone [33] have been shown to suppress AR-Vs protein expression. Meanwhile, antisense oligonucleotides (ASO) specifically for AR can target AR genes and its splice variants have been reported [34]. Additional strategy is to target the transactive domain of AR-Vs by developing small molecular inhibitors or peptidomimetics [35], in hope to suppress their activities by preventing their binding to downstream gene promoter or causing AR-V protein to degrade. However, it is known that many co-activators and co-repressors have been identified to be associated with AR protein. It is likely that this strategy may only provide partial suppression of AR-V activities.
Collectively, AR-Vs are mostly generated by alternative gene splicing in CRPC cells, which offer an additional targeting strategy. Although programmed alternative pre-mRNA splicing is commonly associated with normal cells, more active pre-mRNA splicing or aberrant splicing is often detected in cancer cells [20; 21]. As such, therapeutic intervention targeting the pre-mRNA splicing machinery (spliceosome) has gained considerable attention in recent years [24]. There have been a significant number of explorations to develop spliceosome inhibitors including FR, SSA, pladienolides, and sudemycins [24], Among them, spliceosome inhibitor E7107 has been put to phase I study and shown the desired target effect with dose-dependent pharmacokinetic behavior clinically [36]. These result further highlighted that spliceosome inhibitors are becoming next promising agent to target cancer. Herein, TSTs appeared to be highly potent agents (IC50 in the nM range) to inhibit CRPC cells growth by targeting AR pre-mRNA splicing (Fig. 1 and 4). We confirmed AR-V7 as a major target of TSTs because CRPC cells are highly dependent on AR-V7 for their growth and survival (Fig. 3). Our data further unveiled that the possible mechanism of action of TSTs is to interfere splicing factors binding between U2AF65 and SAP155 and further prevent the binding of splicing complex to 3’ splice site (SS) of AR-V7 pre-mRNA (Fig. 5B and 5D). U2AF has been shown to play a key role in recognizing the alternative 3’-SS of pre-mRNA, For example, in leukemia [27], U2AF35 but not U2AF65 is responsible for exon skipping. In PCa, Liu et al. identified U2AF65 as one of key factors in AR-V7 gene splicing [26]. Indeed, our data indicated that elevated U2AF65 could significantly increase AR-V7 protein expression but not steroidogenesis gene (e.g., AKR1C3) or other house keeping gene (e.g., actin) (Fig. 5C), which may also imply that AR-V gene splicing is highly active in CRPC cells compared with these genes. Nevertheless, U2AF35 appeared not to participate in AR-V7 gene splicing (Fig. S5B and S5C). On the other hand, it is very likely that TSTs can target gene splicing of other genes in CRPC cells. For example, proteins such as CD44 and VEGF with different isoforms derived from alternative splicing[37] in PCa cells could be the potential target(s) of TSTs, which warrants a more comprehensive study in the future.
Based on the mechanism of action of TSTs with respect to the “specificity” of alternative gene splicing unveiled from this study, we also recognize the potential challenge for its side effect once it is delivered systematically. Alternatively, engineering a PCa-specific delivery vehicle could reduce any undesirable side effect. On the other hand, based on its structural functional activities on complex formation of splicing factors, it is possible to design different analogs with high specificity.
In conclusion, our data offer a new therapeutic strategy to target AR-V7 that plays a critical role in not only the refraction to ADT but also the growth and survival of CRPC. Although we only delineated the underlying mechanism of TSTs in intervening AR-V7 gene splicing, we believe TSTs can target other AR-Vs as well as other gene products generated from active alternative splicing in CRPC cells. Thus, TSTs have a great clinical potential to be an effective therapeutic for recurrent CRPC.
Supplementary Material
Novelty & Impact Statements.
AR-V7 is the most characterized AR variants and has been associated with the poor survival of castration resistant prostate cancer (CRPC) patients. Currently, no effective agent can target AR-V7. We unveil the mechanism of action of alternative AR gene splicing and explore potential agent (Thailanstatins, TST) to inhibit AR-7 gene expression. TST appears to be a potent agent in suppressing CRPC cells in vitro and in vivo.
Acknowledgments
We thank Mary Barnes for editorial assistance.
Grand Support: This work was supported by a Pilot Grant (to Y.Q.C. and J.T.H.) from UT Southwestern Medical Center for Translational Medicine funded by NIH’s National Center for Advancing Translational Sciences (5UL1TR001105) and grants from the National Natural Science Foundation of China (NSFC 81202014 to K.J.W and NSFC 81130041 to D.L.H.)
Abbreviations
- TST
Thailanstatin
- AR
Androgen Receptor
- AR-V7
Androgen Receptor Variant 7
- U2AF65
U2 Auxiliary Factor 65
- SAP155
Spliceosome-Associated Protein 155
- SF3B
Splicing Factor 3b.
Footnotes
All the authors don’t have any conflict of interest.
References
- 1.Siegel R, Ma J, Zou Z, Jemal A. Cancer statistics, 2014. CA: A Cancer Journal for Clinicians. 2014;64:9–29. doi: 10.3322/caac.21208. [DOI] [PubMed] [Google Scholar]
- 2.Heinlein CA, Chang C. Androgen Receptor in Prostate Cancer. Endocrine Reviews. 2004;25:276–308. doi: 10.1210/er.2002-0032. [DOI] [PubMed] [Google Scholar]
- 3.Chen Y, Sawyers CL, Scher HI. Targeting the androgen receptor pathway in prostate cancer. Current Opinion in Pharmacology. 2008;8:440–448. doi: 10.1016/j.coph.2008.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Heemers HV, Tindall DJ. Androgen Receptor (AR) Coregulators: A Diversity of Functions Converging on and Regulating the AR Transcriptional Complex. Endocrine Reviews. 2007;28:778–808. doi: 10.1210/er.2007-0019. [DOI] [PubMed] [Google Scholar]
- 5.Dehm SM, Schmidt LJ, Heemers HV, Vessella RL, Tindall DJ. Splicing of a Novel Androgen Receptor Exon Generates a Constitutively Active Androgen Receptor that Mediates Prostate Cancer Therapy Resistance. Cancer Research. 2008;68:5469–5477. doi: 10.1158/0008-5472.CAN-08-0594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chang K, Ercole CE, Sharifi N. Androgen metabolism in prostate cancer: from molecular mechanisms to clinical consequences. British Journal of Cancer. 2014;111:1249–1254. doi: 10.1038/bjc.2014.268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shtivelman E, Beer TM, Evans CP. Molecular pathways and targets in prostate cancer. Oncotarget. 2014;5:7217–59. doi: 10.18632/oncotarget.2406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chang KH, Li R, Papari-Zareei M, Watumull L, Zhao YD, Auchus RJ, Sharifi N. Dihydrotestosterone synthesis bypasses testosterone to drive castration-resistant prostate cancer. Proceedings of the National Academy of Sciences. 2011;108:13728–13733. doi: 10.1073/pnas.1107898108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Harris WP, Mostaghel EA, Nelson PS, Montgomery B. Androgen deprivation therapy: progress in understanding mechanisms of resistance and optimizing androgen depletion. Nature Clinical Practice Urology. 2009;6:76–85. doi: 10.1038/ncpuro1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ryan CJ, Smith MR, Fizazi K, Saad F, Mulders PF, Sternberg CN, Miller K, Logothetis CJ, Shore ND, Small EJ, Carles J, Flaig TW, Taplin ME, Higano CS, de Souza P, de Bono JS, Griffin TW, De Porre P, Yu MK, Park YC, Li J, Kheoh T, Naini V, Molina A, Rathkopf DE. Abiraterone acetate plus prednisone versus placebo plus prednisone in chemotherapy-naive men with metastatic castration-resistant prostate cancer (COU-AA-302): final overall survival analysis of a randomised, double-blind, placebo-controlled phase 3 study. Lancet Oncol. 2015;16:152–60. doi: 10.1016/S1470-2045(14)71205-7. [DOI] [PubMed] [Google Scholar]
- 11.Beer TM, Tombal B. Enzalutamide in metastatic prostate cancer before chemotherapy. N Engl J Med. 2014;371:1755–6. doi: 10.1056/NEJMc1410239. [DOI] [PubMed] [Google Scholar]
- 12.Lu C, Luo J. Decoding the androgen receptor splice variants. Transl Androl Urol. 2013;2:178–186. doi: 10.3978/j.issn.2223-4683.2013.09.08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Thadani-Mulero M, Portella L, Sun S, Sung M, Matov A, Vessella RL, Corey E, Nanus DM, Plymate SR, Giannakakou P. Androgen Receptor Splice Variants Determine Taxane Sensitivity in Prostate Cancer. Cancer Research. 2014;74:2270–2282. doi: 10.1158/0008-5472.CAN-13-2876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Antonarakis ES, Lu C, Wang H, Luber B, Nakazawa M, Roeser JC, Chen Y, Mohammad TA, Chen Y, Fedor HL, Lotan TL, Zheng Q, De Marzo AM, Isaacs JT, Isaacs WB, Nadal R, Paller CJ, Denmeade SR, Carducci MA, Eisenberger MA, Luo J. AR-V7 and resistance to enzalutamide and abiraterone in prostate cancer. N Engl J Med. 2014;371:1028–38. doi: 10.1056/NEJMoa1315815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brand LJ, Dehm SM. Androgen receptor gene rearrangements: new perspectives on prostate cancer progression. Curr Drug Targets. 2013;14:441–9. doi: 10.2174/1389450111314040005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nyquist MD, Li Y, Hwang TH, Manlove LS, Vessella RL, Silverstein KAT, Voytas DF, Dehm SM. TALEN-engineered AR gene rearrangements reveal endocrine uncoupling of androgen receptor in prostate cancer. Proceedings of the National Academy of Sciences. 2013;110:17492–17497. doi: 10.1073/pnas.1308587110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Robinson D, Van Allen EM, Wu YM, Schultz N, Lonigro RJ, Mosquera JM, Montgomery B, Taplin ME, Pritchard CC, Attard G, Beltran H, Abida W, Bradley RK, Vinson J, Cao X, Vats P, Kunju LP, Hussain M, Feng FY, Tomlins SA, Cooney KA, Smith DC, Brennan C, Siddiqui J, Mehra R, Chen Y, Rathkopf DE, Morris MJ, Solomon SB, Durack JC, Reuter VE, Gopalan A, Gao J, Loda M, Lis RT, Bowden M, Balk SP, Gaviola G, Sougnez C, Gupta M, Yu EY, Mostaghel EA, Cheng HH, Mulcahy H, True LD, Plymate SR, Dvinge H, Ferraldeschi R, Flohr P, Miranda S, Zafeiriou Z, Tunariu N, Mateo J, Perez-Lopez R, Demichelis F, Robinson BD, Schiffman M, Nanus DM, Tagawa ST, Sigaras A, Eng KW, Elemento O, Sboner A, Heath EI, Scher HI, Pienta KJ, Kantoff P, de Bono JS, Rubin MA, Nelson PS, Garraway LA, Sawyers CL, Chinnaiyan AM. Integrative clinical genomics of advanced prostate cancer. Cell. 2015;161:1215–28. doi: 10.1016/j.cell.2015.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dehm SM, Tindall DJ. Alternatively spliced androgen receptor variants. Endocrine Related Cancer. 2011;18:R183–R196. doi: 10.1530/ERC-11-0141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pan Q, Shai O, Lee LJ, Frey BJ, Blencowe BJ. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nature Genetics. 2008;40:1413–1415. doi: 10.1038/ng.259. [DOI] [PubMed] [Google Scholar]
- 20.Mills JD, Janitz M. Alternative splicing of mRNA in the molecular pathology of neurodegenerative diseases. Neurobiology of Aging. 2012;33:1012.e11–1012.e24. doi: 10.1016/j.neurobiolaging.2011.10.030. [DOI] [PubMed] [Google Scholar]
- 21.Dehm SM. mRNA Splicing Variants: Exploiting Modularity to Outwit Cancer Therapy. Cancer Research. 2013;73:5309–5314. doi: 10.1158/0008-5472.CAN-13-0444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu X, Biswas S, Berg MG, Antapli CM, Xie F, Wang Q, Tang M, Tang G, Zhang L, Dreyfuss G, Cheng Y. Genomics-Guided Discovery of Thailanstatins A, B, and C As Pre-mRNA Splicing Inhibitors and Antiproliferative Agents fromBurkholderia thailandensis MSMB43. Journal of Natural Products. 2013;76:685–693. doi: 10.1021/np300913h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu Xiangyang, Sbay HZ. Improved production of cytotoxic thailanstatins A and D through metabolic engineering of Burkholderia thailandensis MSMB43 and pilot scale fermentation. Synthetic and Systems Biotechnology. 2016 doi: 10.1016/j.synbio.2016.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kaida D, Motoyoshi H, Tashiro E, Nojima T, Hagiwara M, Ishigami K, Watanabe H, Kitahara T, Yoshida T, Nakajima H, Tani T, Horinouchi S, Yoshida M. Spliceostatin A targets SF3b and inhibits both splicing and nuclear retention of pre-mRNA. Nature Chemical Biology. 2007;3:576–583. doi: 10.1038/nchembio.2007.18. [DOI] [PubMed] [Google Scholar]
- 25.Liu C, Lou W, Zhu Y, Nadiminty N, Schwartz CT, Evans CP, Gao AC. Niclosamide Inhibits Androgen Receptor Variants Expression and Overcomes Enzalutamide Resistance in Castration-Resistant Prostate Cancer. Clinical Cancer Research. 2014;20:3198–3210. doi: 10.1158/1078-0432.CCR-13-3296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu LL, Xie N, Sun S, Plymate S, Mostaghel E, Dong X. Mechanisms of the androgen receptor splicing in prostate cancer cells. Oncogene. 2014;33:3140–50. doi: 10.1038/onc.2013.284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shao C, Yang B, Wu T, Huang J, Tang P, Zhou Y, Zhou J, Qiu J, Jiang L, Li H, Chen G, Sun H, Zhang Y, Denise A, Zhang D, Fu X. Mechanisms for U2AF to define 3′ splice sites and regulate alternative splicing in the human genome. Nature Structural & Molecular Biology. 2014;21:997–1005. doi: 10.1038/nsmb.2906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kralovicova J, Knut M, Cross NCP, Vorechovsky I. Identification of U2AF(35)-dependent exons by RNA-Seq reveals a link between 3' splice-site organization and activity of U2AF-related proteins. Nucleic Acids Research. 2015;43:3747–3763. doi: 10.1093/nar/gkv194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Myung J, Banuelos CA, Fernandez JG, Mawji NR, Wang J, Tien AH, Yang YC, Tavakoli I, Haile S, Watt K, McEwan IJ, Plymate S, Andersen RJ, Sadar MD. An androgen receptor N-terminal domain antagonist for treating prostate cancer. Journal of Clinical Investigation. 2013;123:2948–2960. doi: 10.1172/JCI66398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Antonarakis ES, Lu C, Luber B, Wang H, Chen Y, Nakazawa M, Nadal R, Paller CJ, Denmeade SR, Carducci MA, Eisenberger MA, Luo J. Androgen Receptor Splice Variant 7 and Efficacy of Taxane Chemotherapy in Patients With Metastatic Castration-Resistant Prostate Cancer. JAMA Oncology. 2015;1:582. doi: 10.1001/jamaoncol.2015.1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yamashita S, Lai KP, Chuang KL, Xu D, Miyamoto H, Tochigi T, Pang ST, Li L, Arai Y, Kung HJ, Yeh S, Chang C. ASC-J9 suppresses castration-resistant prostate cancer growth through degradation of full-length and splice variant androgen receptors. Neoplasia. 2012;14:74–83. doi: 10.1593/neo.111436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Asangani IA, Dommeti VL, Wang X, Malik R, Cieslik M, Yang R, Escara-Wilke J, Wilder-Romans K, Dhanireddy S, Engelke C, Iyer MK, Jing X, Wu YM, Cao X, Qin ZS, Wang S, Feng FY, Chinnaiyan AM. Therapeutic targeting of BET bromodomain proteins in castration-resistant prostate cancer. Nature. 2014;510:278–82. doi: 10.1038/nature13229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kwegyir-Afful AK, Ramalingam S, Purushottamachar P, Ramamurthy VP, Njar VC. Galeterone and VNPT55 induce proteasomal degradation of AR/AR-V7, induce significant apoptosis via cytochrome c release and suppress growth of castration resistant prostate cancer xenografts in vivo. Oncotarget. 2015;6:27440–60. doi: 10.18632/oncotarget.4578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yamamoto Y, Loriot Y, Beraldi E, Zhang F, Wyatt AW, Al NN, Mo F, Zhou T, Kim Y, Monia BP, MacLeod AR, Fazli L, Wang Y, Collins CC, Zoubeidi A, Gleave M. Generation 2.5 antisense oligonucleotides targeting the androgen receptor and its splice variants suppress enzalutamide-resistant prostate cancer cell growth. Clin Cancer Res. 2015;21:1675–87. doi: 10.1158/1078-0432.CCR-14-1108. [DOI] [PubMed] [Google Scholar]
- 35.Ravindranathan P, Lee T, Yang L, Centenera MM, Butler L, Tilley WD, Hsieh J, Ahn J, Raj GV. Peptidomimetic targeting of critical androgen receptor–coregulator interactions in prostate cancer. Nature Communications. 2013;4:1923. doi: 10.1038/ncomms2912. [DOI] [PubMed] [Google Scholar]
- 36.Eskens FALM, Ramos FJ, Burger H, O'Brien JP, Piera A, de Jonge MJA, Mizui Y, Wiemer EAC, Carreras MJ, Baselga J, Tabernero J. Phase I Pharmacokinetic and Pharmacodynamic Study of the First-in-Class Spliceosome Inhibitor E7107 in Patients with Advanced Solid Tumors. Clinical Cancer Research. 2013;19:6296–6304. doi: 10.1158/1078-0432.CCR-13-0485. [DOI] [PubMed] [Google Scholar]
- 37.David CJ, Manley JL. Alternative pre-mRNA splicing regulation in cancer: pathways and programs unhinged. Genes & Development. 2010;24:2343–2364. doi: 10.1101/gad.1973010. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.