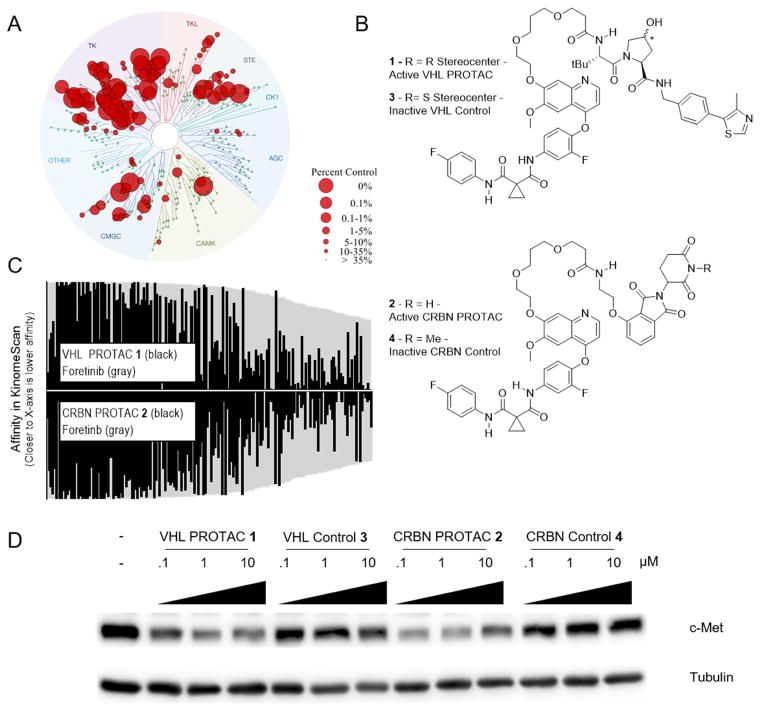
Figure 1. Foretinib PROTACs bind many kinases and potently degrade c-Met.
(A) Foretinib binds to 133 kinases with a Percent of Control less than 35 as measured by a competitive binding assay. (B) Structures of the foretinib-based PROTACs used in this study. Top is the VHL-recruiting PROTAC compound 1. The stereocenter of the hydroxyproline VHL-binding element is R for the active PROTAC while it is S for the inactive control compound 3. Bottom, the Cereblon-recruiting PROTAC compound 2. The nitrogen atom of the glutarimide ring is non-methylated in the active PROTAC, while in the inactive control compound 4 it is methylated. (C) The affinity for most kinases changes upon addition of the linker and E3-ligase recruiting element. Along the X-axis are the 133 kinases that are considered hits for foretinib (i.e. having a percent of control value of 35 or less in KinomeScan data) sorted in order of decreasing affinity for foretinib. The values for foretinib are plotted in grey on both the positive and negative Y-axes. On the positive Y-axis, the values for compound 1 are plotted in black, while the negative X-axis has the values for compound 2. See Supplemental Table 1 and Figure S1 for full KinomeScan data sets. (D) MDA-MB-231 cells were treated with the indicated concentrations of compounds 1–4 for 24 hours, and c-Met protein levels were analyzed by immunoblot.