Figure 3. Endothelial CXCR4 reduces vascular permeability through WNT signaling.
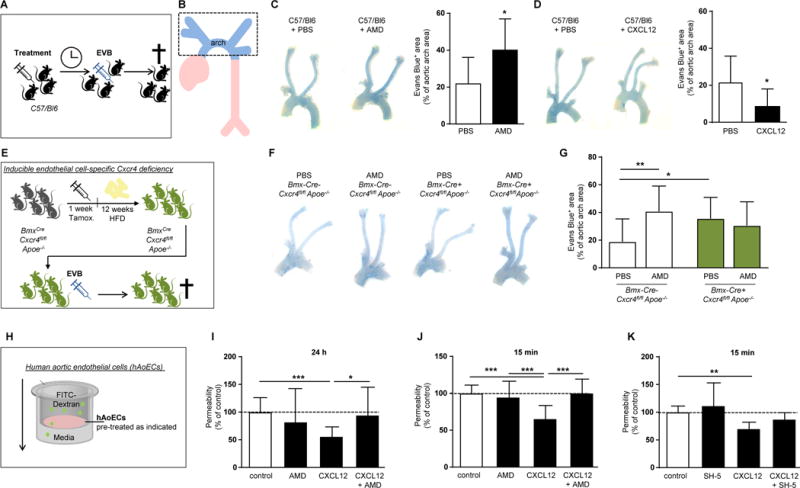
(A, B) Experimental scheme. (C) Histamine-induced (10 µg i.v. for 10 min) vascular permeability to Evans blue in C57/Bl6 mice pretreated with the CXCR4 antagonist AMD3465 (125 µg/mouse) or vehicle (PBS) for 16 hours, as indicated. (n=8-13; Student’s t-test with Welch correction). Representative images are shown (left panel). (D) Histamine-induced (10 µg histamine i.v. for 10 min) vascular permeability to Evans blue in C57/Bl6 mice pretreated for 4 hours with CXCL12 (3 µg) or vehicle (PBS), as indicated (n=8-13; Student’s t-test with Welch correction). Representative images are shown (left panel). (E) Experimental scheme. (F, G) Evans blue extravasation in the aortic arch of BmxCre-Cxcr4fl/flApoe−/− and BmxCre+ Cxcr4fl/flApoe−/− mice after 12 weeks of HFD and treated with the CXCR4 antagonist AMD3465 (125 µg/mouse) or vehicle (PBS) for 12 hours, as indicated. (n=9-21; 2-way ANOVA with Bonferroni post-test). Representative images are shown (left panel). (H) Experimental scheme. (I,J) Permeability of hAoECs to FITC-Dextran after stimulation with CXCL12 (100 ng/ml) for 24 h (I) or 15 min (J). Where indicated, cells were pretreated with the CXCR4 antagonist AMD3100 (I) or AMD3465 (J) at 1 µg/ml for 1 h before CXCL12 stimulation. Data in (I) represent n=17-20 wells from 7 independent experiments (Kruskal-Wallis test with Dunn’s post-test). Data in (J) represent n=11-19 wells from 6 independent experiments (1-way ANOVA with Tukey’s multiple comparison test). (K) Permeability of hAoECs to FITC-Dextran after stimulation for 15 min with CXCL12 (100 ng/ml) and pretreated with the Akt inhibitor SH-5 (20 nM) for 30 min, as indicated (n=6-8 wells from 4 independent experiments; Kruskal-Wallis test with Dunn’s post-test). (C-K) Graphs represent mean mean±SD, *P<0.05; **P<0.01; ***P<0.001.
