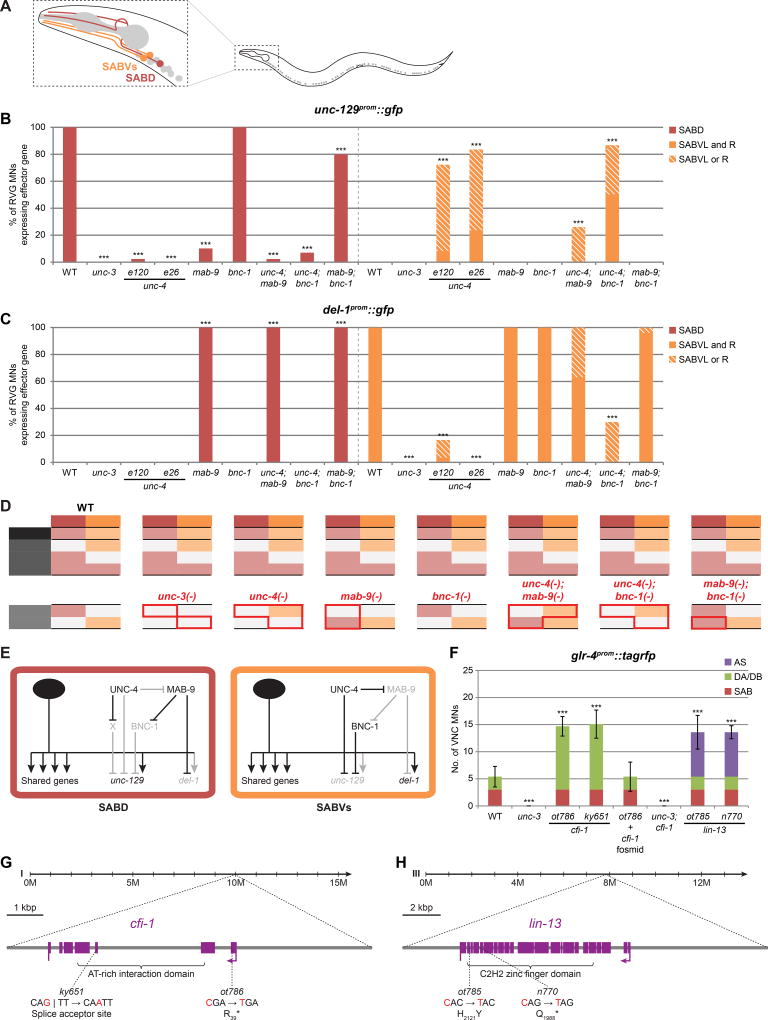
Figure 5. A repressor regulatory logic also operates in head SAB motor neurons.
A: SAB MNs in the RVG and their relative positions and morphology.
B: unc-129 expression is specific to SABD and is unc-3-dependent. This expression is lost in mab-9 mutants in a bnc-1-dependent manner, indicating that MAB-9 represses bnc-1 which represses unc-129. Similarly, unc-129 expression in SABD is lost in unc-4 mutants. This effect is likely also indirect with UNC-4 possibly repressing an unidentified repressor of unc-129 (note that e26 disrupts UNC-4 interaction with the Groucho-like corepressor UNC-37). This repressor is not bnc-1 as the bnc-1 mutant phenotype is not epistatic to that of unc-4. On the other hand, unc-129 is derepressed in SABVs (albeit incompletely) in unc-4 mutants in a mab-9-dependent manner, indicating that UNC-4 represses mab-9. As in SABD, mab-9 is likely repressing bnc-1 which represses unc-129. This predicts that bnc-1 mutants would phenocopy those of unc-4 – which is not the case. UNC-4 might independently to, and redundantly with bnc-1 be repressing unc-129 in SABVs. Fischer’s exact tests were performed for all mutants compared to WT; ***p < 0.001; n ≥ 25.
C: del-1 expression is specific to SABVs and is unc-3-dependent. In mab-9 mutants, del-1 is derepressed in SABD. In unc-4 mutants, del-1 expression is lost in SABVs in a mab-9-dependent manner, indicating that UNC-4 represses mab-9 which represses del-1. Fischer’s exact tests were performed for all mutants compared to WT; ***p < 0.001; n ≥ 20.
D: Effect of repressor mutants on class-specific effector genes in SAB MNs. Note expression pattern of repressor genes (see Fig.S4A). Red bounding lines highlight changes in effector gene expression in repressor mutants compared to WT.
E: Genetic model depicting the interactions of repressors on class-specific effector genes in SAB MNs.
F: glr-4 is expressed in SAB MNs in an unc-3-dependent manner, but expression is largely absent in VNC MNs. In cfi-1(ot786), glr-4 is derepressed in DA/DB MNs (phenocopied by ky651) in an unc-3-dependent manner. This phenotype is rescued by a fosmid containing the cfi-1 locus. Additionally, in lin-13(ot785), glr-4 is derepressed in AS MNs (phenocopied by n770). Error bars show SD; unpaired t-tests were performed for all mutants compared to WT; ***p < 0.001; n ≥ 13. See Fig.S4C,D for images.
G: cfi-1 gene locus.
H: lin-13 gene locus. Not shown are four regulatory mutations in ot785.