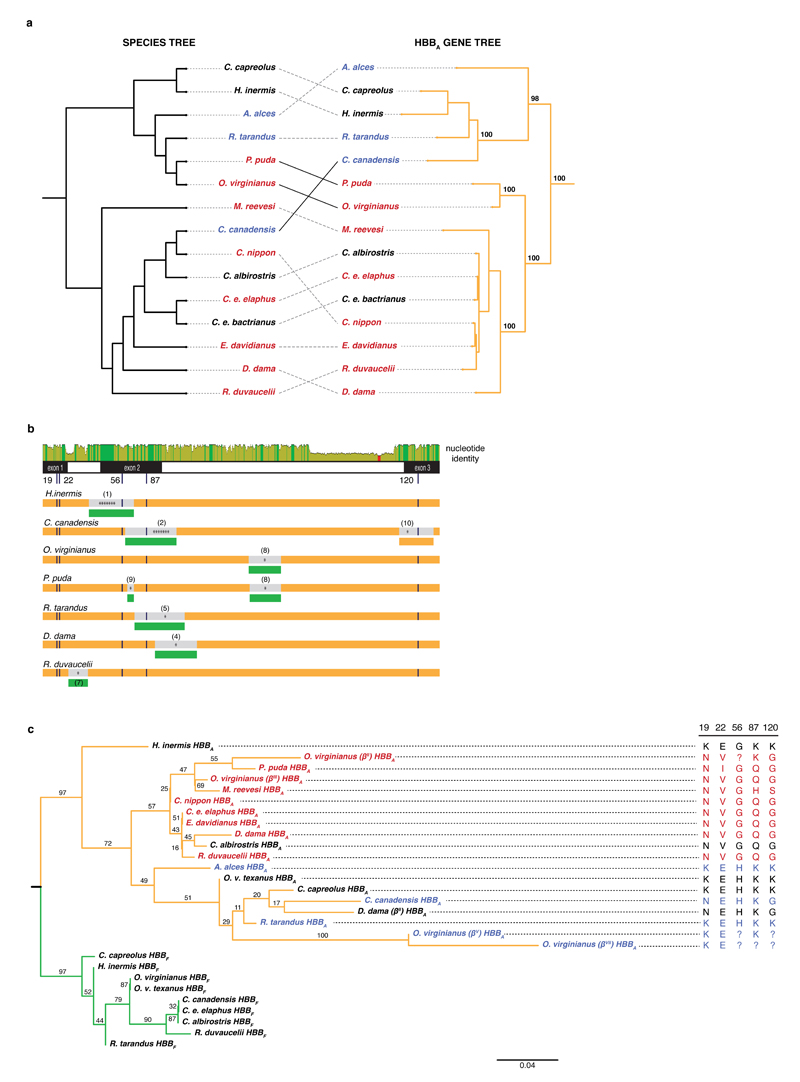
Fig. 3. Evidence for incomplete lineage sorting, gene conversion, and a trans-species polymorphism in the evolutionary history of deer HBBA.
a, Discordances between the maximum likelihood HBBA gene tree and the species tree. Topological differences that violate the principal division into New World deer (NWD, Capreolinae) and Old World deer (OWD, Cervinae) are highlighted by solid black lines. Bootstrap values (% out of 1000 bootstrap replicates) are highlighted for salient nodes. b, Gene conversion and/or introgression. The top panel illustrates nucleotide identity between HBBA and HBBF orthologs (green: 100%, yellow: 30-100%, red: <30% identity). The low-identity segment towards the end of intron 2 marks repeat elements present in all adult but absent from all foetal sequences. Below, predicted recombination events affecting HBBA genes (orange), with either an adult ortholog (orange) or a foetal HBBF paralog (green) as the predicted source, suggestive of introgression or gene conversion, respectively. The number of asterisks indicates how many detection methods (out of a maximum of seven) predicted a given event (see Methods). Details for individual events (numbered in parentheses) are given in Supplementary Figure 8. c, Maximum likelihood protein tree of adult (orange) and foetal (green) β-globin. Alternate non-sickling D. dama (II) and O. virginianus (V, VII) alleles group with non-sickling species (coloured as in Fig. 1). Amino acid identity at key sites is shown on the right. ?: amino acid unresolved in primary source.