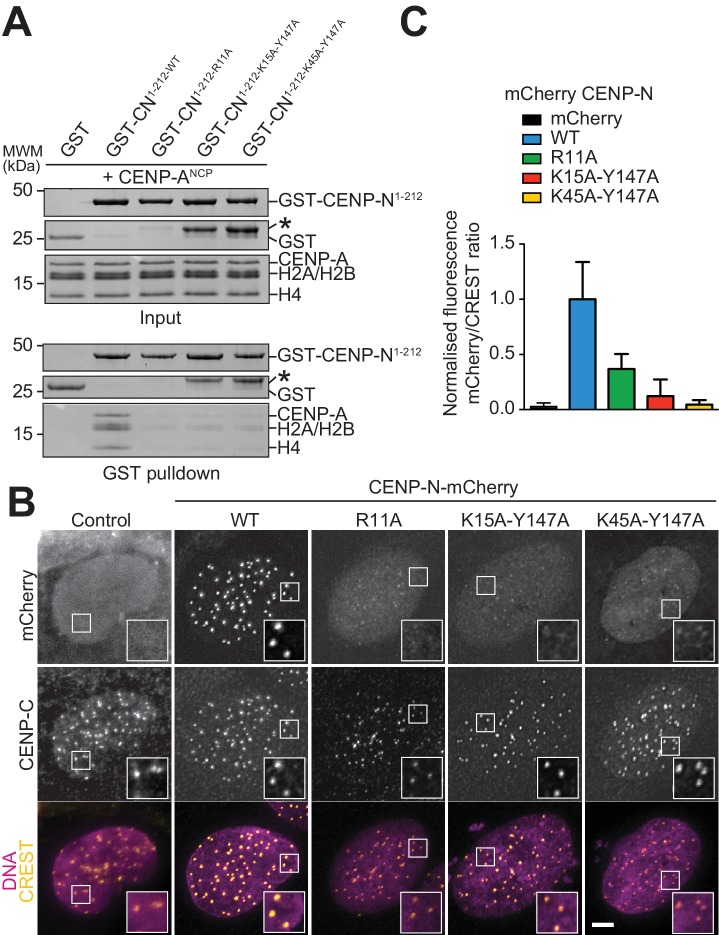
Figure 4. Validation of the CENP-N:CENP-ANCP complex.
(A) In vitro binding assay probing the interaction of GST-CENP-N1-212 immobilized on solid phase with CENP-ANCP. (B) Fluorescence microscopy analysis comparing localization at human kinetochores (U2OS osteosarcoma cells) of a wild-type CENP-N-mCherry fluorescent reporter and of its mutant variants. (C) Quantification of localization of the mCherry constructs in B normalized to CREST. The same concentrations of transiently transfected plasmids were compared. Error bars represent SD.