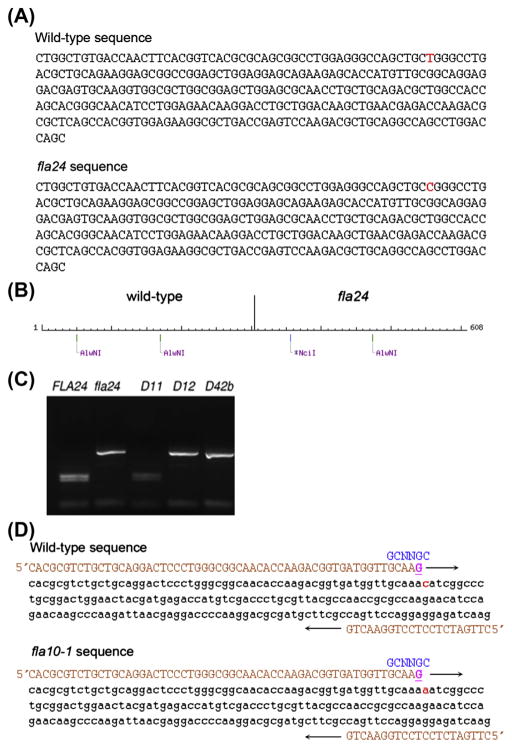
FIGURE 2. Mapping and dCAPS design.
(A). Genomic sequence around wild-type and fla24 mutation. The red nucleotide indicates the mutant SNP (T to C). (B) NEBcutter (Vincze, Posfai, & Roberts, 2003) was used to cut the wild-type and the fla24 sequence. The wild-type sequence is cut twice by AlwNI and the fla24 mutant sequence is cut only once by AlwNI and by NciI. Enzymes that cut an odd number of times in the combined wild-type and mutant sequence are desirable (C). PCR products cut by AlwNI. Wild-type and the true revertant D11 are cut twice by AlwNI. (Reproduced from (Lin, Nauman, et al., 2013).) (D) The change in fla10-1, which has an N to K change, does not have a restriction enzyme site difference between wild-type and the mutant. A dCAPS (derived cleaved amplified polymorphic sequence) marker was designed (Neff et al., 2002). The primers (in orange (light gray in print versions)) introduce a nucleotide change that generates a restriction enzyme recognition site. The forward primer (in orange (light gray in print versions)) introduces a G (underlined in magenta (light gray in print versions)) that creates a restriction site (in blue (black in print versions)) in the wild-type sequence but not in the mutant sequence. The mutant nucleotide is shown in red (light gray in print versions).