Fig. 10. The quorum sensing regulator HapR is a vieSAB repressor.
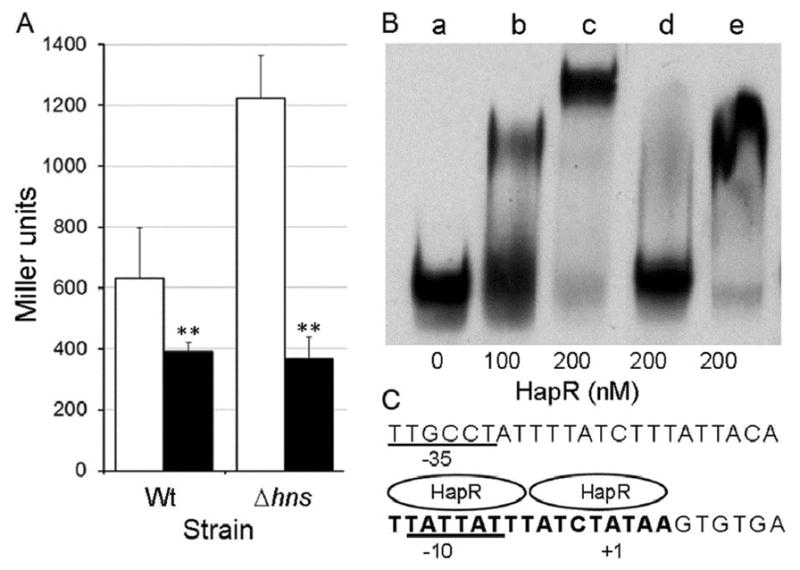
A. Strain O395ΔlacZ (Wt) and O395ΔlacZΔhns (Δhns) containing a vieSAB-lacZ fusion were transformed with parent plasmid pACY184 (□. open bars) or pC1.1 encoding HapR (■, filled bars). The strains were grown in LB and β-galactosidase activity (Miller units) was measured as an indicator of promoter activity. The error bars denote the standard deviation of three independent transformants. Symbols: ** p < 0.01 (unpaired T test). B. A DIG-labeled vieSAB promoter fragment spanning nucleotide −198 to +70 relative to the TSS was treated with increasing concentration of pure HapR (lanes b and c), 200 nM HapR in the presence of a 200-fold excess of unlabeled competitor vieSAB DNA (lane d) or 200 nM HapR in the presence of a 200-fold excess of unlabeled DNA upstream aphA lacking the HapR binding site (amplified with primers AphA51 and AphA149). C. Predicted HapR binding site (bold font) at the vieSAB promoter relative to the TSS (+1 nucleotide) and −10/−35 elements.
