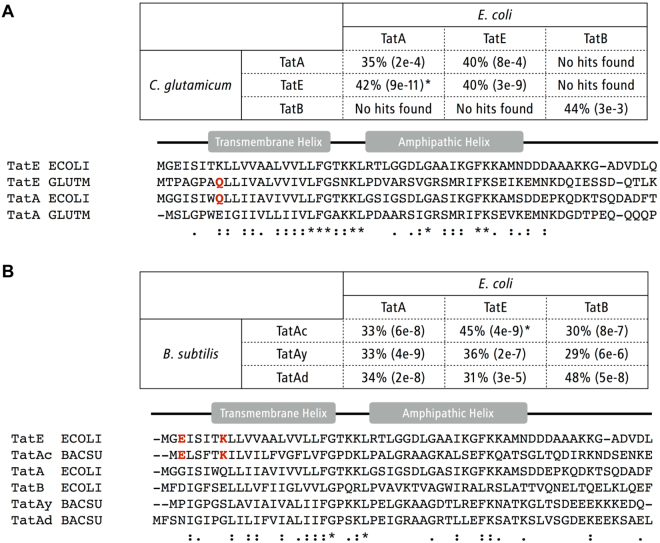
Figure 5.
TatE paralogs in Gram-positive bacteria. The degree of sequence similarity, indicated by the percent of identity and the expect e-value (which decreases with the lower chance of random match) between Tat proteins from Escherichia coli and Corynebacterium glutamicum (A) and Bacillus subtilis (B), respectively, were calculated using NCBI BLAST (upper panels). (A) The closest homologue of C. glutamicum TatE is TatA from E. coli, both featuring non-charged N-termini as revealed in the multiple sequence alignment (lower panel). (B) In B. subtilis, TatAc is most similar to TatE of E coli, as shown by the common ExxxxK motif. Sequence source: C. glutamicum TatA (Q8NQE4), TatE (Q2MGW8), TatB (Q8NRD0); B. subtilis TatAy (O05522), TatAc (O31804), TatAd (O31467).