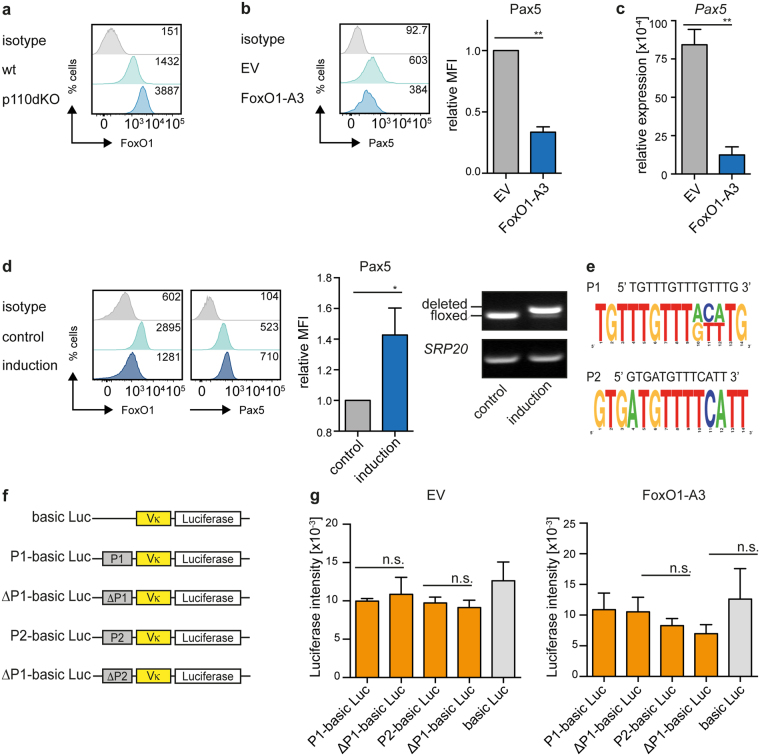
Figure 5.
PI3K regulates Pax5 via FoxO1. (a) Cells from bm-derived wt and p110dKO pre-B cell lines were analyzed by intracellular FACS for FoxO1. (b) Cells from a bm-derived wt pre-B cell line were transduced with a constitutively active form of FoxO1 (FoxO1-A3) or an EV control. Expression of Pax5 was assessed by intracellular FACS at day 2 after transduction in at least 3 independent experiments. Statistical significance was calculated using the t-Test. (c) Total RNA of cells from Fig. 5b was isolated to analyze transcript levels of Pax5. Hprt and Pax5 mRNA-levels were detected by qRT-PCR using the SYBR-Green detection method. Results are shown as mean ± SD of 2 independent experiments, run as duplicates. Statistical significance was calculated using the t-Test. (d) Cells from a FoxO1fl/fl bm-derived pre-B cell line were transduced with Cre-ERT2 or -ERT2. 4-OHT was applied to activate the Cre-ERT2 (induction), treatment with EtOH served as control. Expression levels of FoxO1 and Pax5 were analyzed by intracellular flow cytometry (left panel). The MFI of Pax5 upon deletion of FoxO1 was quantified (bar diagram). Statistical significance was calculated using the t-test. Deletion of the floxed FoxO1 allele was confirmed by PCR, SRP20 served as loading control (right). For original full-length gel pictures see Fig. S2d. Depicted data are representative of at least 3 independent experiments. (e) Sequences of two FoxO1-binding motifs identified by the Encode Genome Project. Site 1 (P1) is located in intron 5, site 2 (P2) in intron 9 of the murine Pax5 gene. P1 and P2 are highly conserved between man and mouse. Graphs were prepared using the WebLogo software. (f) Schematic overview of the luciferase expression vector harboring a Vκ21 promoter and a 1 kb fragment containing or lacking the potential FoxO1 binding sites shown in Fig. 5e. (g) WEHI cells were electroporated to introduce the empty vector (Vκ + Luciferase) or the indicated constructs containing or lacking the potential FoxO1 binding sites. Expression of luciferase was equalized to the rLUC(Renilla) expression in each sample and WEHI cells were cotransfected either with EV (CMV-EV) or with a vector encoding FoxO1-A3 (CMV-FoxO1-A3). Data are representative of 3 (EV) and 4 (FoxO1-A3) independent experiments and luciferase expression was determined using the Dual-Luciferase Reporter Assay System (Promega).