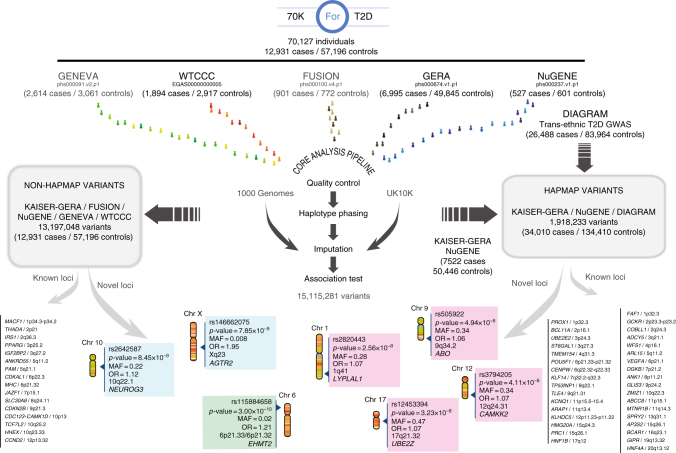
Fig. 1.
Discovery and replication strategy. Publicly available GWAS datasets representing a total of 12,931 cases and 57,196 controls (70KforT2D) were first quality controlled, phased, and imputed, using 1000G and UK10K separately. For those variants that were present in the DIAGRAM trans-ethnic meta-analysis, we used the summary statistics to meta-analyze our results with the cohorts that had no overlap with any of the cohorts included in the DIAGRAM trans-ethnic meta-analysis. With this first meta-analysis, we discovered four novel loci (within magenta panels). For the rest of the variants, we meta-analyzed all the 70KforT2D data sets, which resulted in two novel loci (in blue panels). All the variants that were coding and showed a p-value of ≤ 1 × 10−4 were tested for replication by interrogating the summary statistics in the Type 2 Diabetes Knowledge Portal (T2D Portal) (http://www.type2diabetesgenetics.org/). This uncovered a novel low-frequency variant in the EHMT2 gene (highlighted with a green panel)