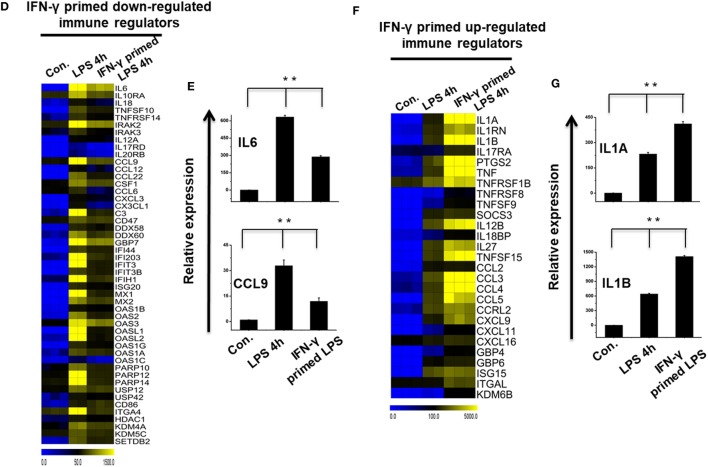
Figure 3.
Comparison of only lipopolysaccharide (LPS)-inducible and interferon-g (IFN-γ)-primed LPS-inducible transcriptional datasets. (A,B) Heat map representation of the transcripts that were uniquely upregulated in LPS-inducible and IFN-γ-primed LPS-inducible bone marrow-derived macrophages (BMDMs), respectively. (C) Gene set enrichment analysis (GSEA) results showing C5 gene ontology (GO) terms collections of the GSEA molecular signatures database (MSigDB) v6.1 in LPS-inducible and IFN-γ-primed LPS-inducible upregulated inflammatory genes in BMDMs. The top GO terms are ranked by the NES. (D,F) Heat map representation of the positive regulators of inflammatory transcripts [cytokines, chemokines, and interferon regulated genes (IRGs)] that were either down or upregulated in IFN-γ-primed LPS-inducible BMDMs compared with only LPS-inducible BMDMs, respectively. (E,G) Validation of IFN-γ-primed LPS-inducible down or upregulated genes compared with only LPS-inducible BMDMs, respectively. Gene expression was normalized to the glyceraldehyde-3-phosphate dehydrogenase transcript levels. **P < 0.001 compared with the control. The data represent three independent biological experiments.