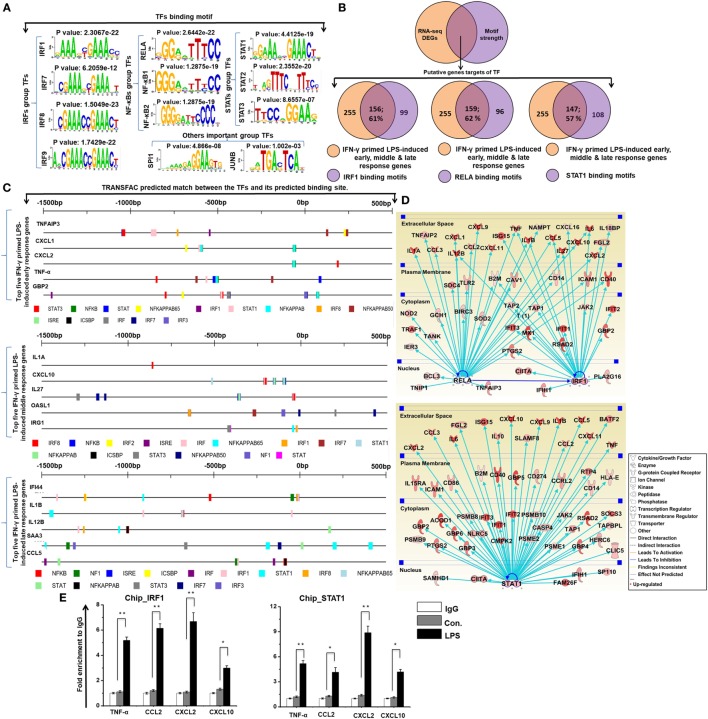
Figure 5.
Transcription factor (TFBSs) analysis within the promoters of common early, middle, and late upregulated genes in interferon-g (IFN-γ)-primed lipopolysaccharide (LPS)-inducible bone marrow-derived macrophages (BMDMs). (A) Patterns of TF motif enrichment within the promoters of the common early, middle and late upregulated genes in IFN-γ-primed LPS-inducible BMDMs (B) Venn diagrams of common early, middle and late upregulated genes associated with interferon regulatory factor-1 (IRF1), RELA, and signal transducer and activator of transcription 1 (STAT1) in BMDMs. (C) TRANSFAC predicted match showing the predicted TFs and their predicted binding sites for the top five early, middle and late upregulated genes in IFN-γ-primed LPS-inducible BMDMs. (D) The activity of highly connected positive regulators of the inflammatory genes IRF1, RELA, and STAT1 led to the activation of this network, as assessed using the ingenuity pathway analysis molecule activity predictor in IFN-γ-primed LPS-inducible BMDMs. (E) Chromatin Immunoprecipitation (ChIP) assay to determine the binding of IRF1, and STAT1 to target genes. The ChIP-enriched samples were subjected to quantitative PCR with selected genes promoters. The graphs represent the mean fold values of enrichment relative to IgG control from three independent experiments. *P ≤ 0.01 and **P ≤ 0.001 compared with the control.