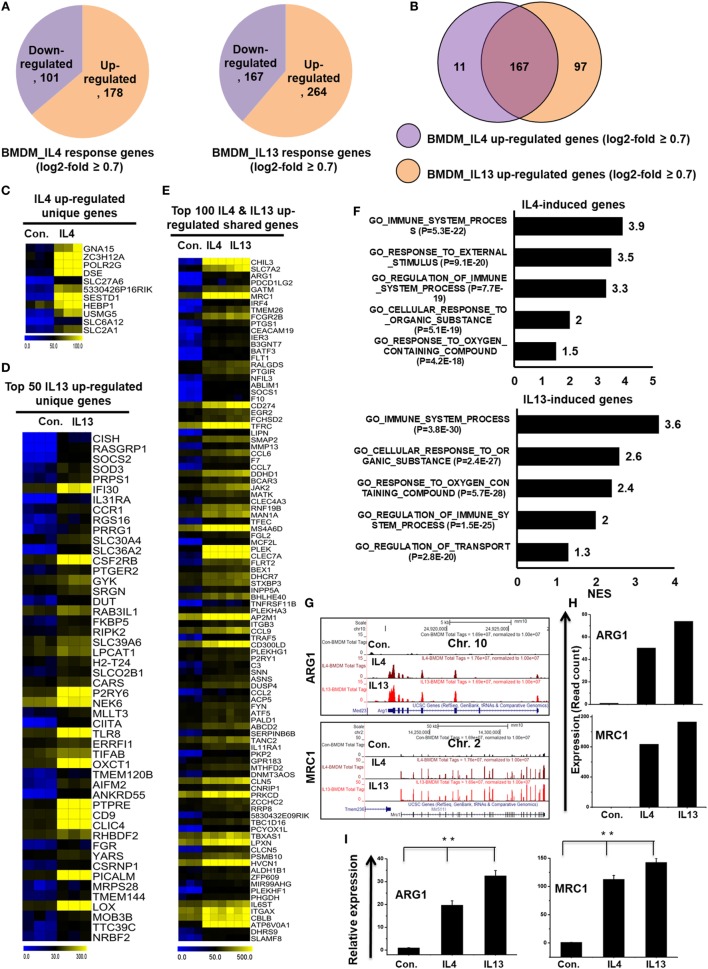
Figure 6.
Differences in transcriptomic profiles between interleukin-4 (IL)-4 and IL-13-treated bone marrow-derived macrophages (BMDMs). (A) Pie chart displaying the number of up or downregulated genes in IL-4 and IL-13 treated BMDMs. (B) The area of overlap indicates the number of unique or shared upregulated genes in IL-4 and IL-13-treated BMDMs. (C,D) Heat map representation of the transcripts that were uniquely upregulated in IL-4 and IL-13-treated BMDMs, respectively. (E) Heat map representation depicting transcripts that were commonly expressed in IL-4 and IL-13-treated BMDMs. (F) Gene set enrichment analysis results of the functional annotations that were associated with the upregulated genes in IL-4 and IL-13-treated BMDMs. (G) UCSC Browser images representing the normalized RNA-seq read density in commonly expressed M2-associated genes between IL-4 and IL-13-treated BMDMs. (H) The transcript abundance (in read count) was evaluated using RNA-seq for commonly expressed M2-associated genes between IL-4 and IL-13-treated BMDMs. (I) Validation of commonly upregulated M2-associated genes in IL-4 and IL-13-treated BMDMs. Gene expression was normalized to glyceraldehyde-3-phosphate dehydrogenase transcript levels. **P < 0.001 compared with the control. The data represent three independent biological experiments.