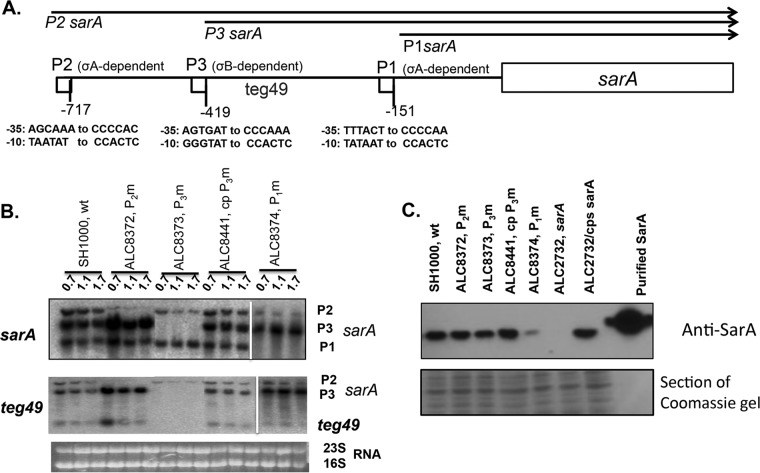
FIG 1.
Expression of the sarA transcripts and teg49 sRNA from sarA promoter mutants at various phases of growth in S. aureus. (A) Schematic representation of the sarA locus showing various transcripts (sarA P2, sarA P3, and sarA P1) that originate from the P2, P3, and P1 promoters (arrows), respectively. The sarA open reading frame is indicated by a box, and the promoters are indicated by small boxes. The numbers of the promoter locations are based on the start codon (ATG) of the sarA gene. (B) Northern blot analysis of total RNA isolated from the wild-type SH1000 and the various isogenic promoter mutants at various phases of growth recorded as OD600 values (OD600 values of 0.7, 1.1, and 1.5 represent mid-log, late log, and early stationary phases, respectively). In all the gels, 10 μg of total cellular RNA was loaded onto each lane, and the blots were probed with a 375-bp sarA DNA probe containing the sarA ORF (top) or a 180-bp DNA fragment containing teg49 (middle). The 16S and 23S rRNAs of the ethidium bromide-stained gel used for blotting is also shown as a loading control at the bottom. The spliced images were taken from different regions of the same image. (C) Western blot analysis for SarA, with anti-SarA antibody, of the wild type, its isogenic promoter mutants, the sarA mutant, and a complemented mutant. Equivalent amounts of extracts (10 μg) from the late exponential phase of growth (OD600 ≈ 1.1) were used to detect SarA protein. (Bottom) A Coomassie blue-stained duplicate-run gel used for blotting is shown as a loading control.