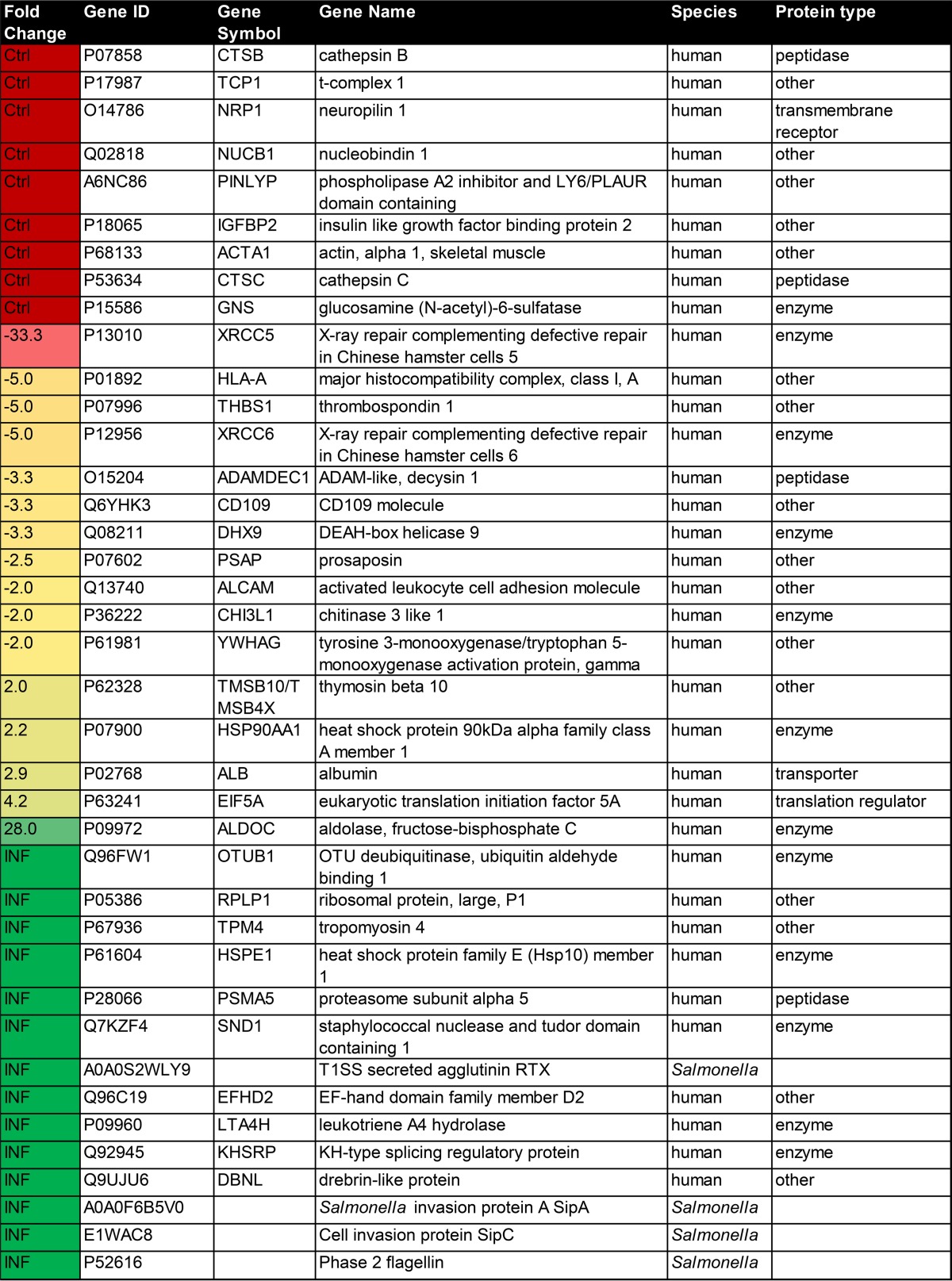
TABLE 1.
Extracellular proteins of THP-1 macrophages with abundance affected by S. Typhimurium infectiona
Fold change indicates up- or downregulation of extracellular proteins from infected cells in comparison to uninfected cells. UniProt ID, gene (protein) name, and species are shown. Gene symbol and protein types are exported from the IPA software (if known). Additional information is available in Table S1 in the supplemental material. The minimum protein identification probability for shown proteins is 95%. The P values were calculated using the Fisher exact test and spectral counts of two independent biological replicates, where a minimum P value was 0.05. SEQUEST identifications required delta Cn scores of greater than 0.2 and XCorr scores of greater than 1.2, 1.9, 2.3, and 2.6 for singly, doubly, triply, and quadruply charged peptides, respectively. The reported peptide FDR was 0.03%, and the protein FDR was 0.2%. Proteins identified only in control or only in infected samples are indicated as Ctrl and INF, respectively.