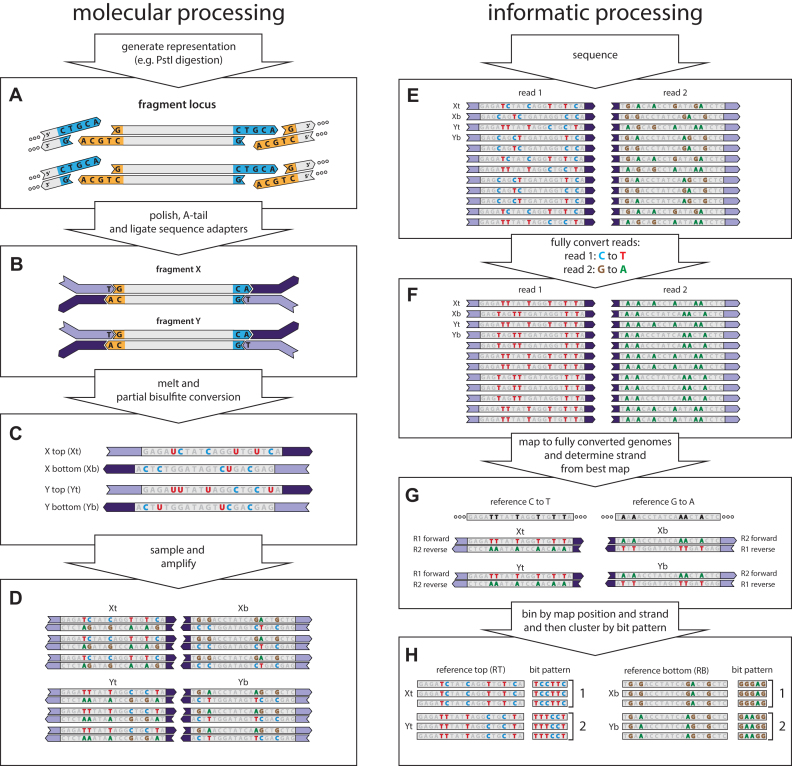
Figure 1.
muSeq applied to a genomic representation. We demonstrate the application of muSeq to a genomic representation by tracing two fragments (X and Y) from a single genomic locus through the molecular and informatics processing steps. The full experiment comprises 160,000 genomic loci and about 100 fragments per locus. (A) We first generate a representation from genomic DNA using PstI. (B) The ends of restriction fragments are polished, T-tailed, and then the fragments are ligated to bisulfite-resistant sequence adapters. (C) The templates are then melted and subjected to partial bisulfite conversion (Materials and Methods). The conversion process randomly deaminates unmethylated cytosines (blue C), converting some proportion to uracil (red U). Each double-stranded molecule results in two templates, one from each strand. (D) We then sample the converted templates and PCR amplify to generate a sequencing library. During amplification uracil is copied as thymine (red T). (E) The library is then paired-end sequenced on an Illumina machine. Because of asymmetries in the sequencing primers, read one shows C to T conversion while read 2 shows G to A conversions. We then fully convert the reads (F) and map them to two fully converted genomes (G): one where every C in the reference is converted to a T and one where every G is converted to an A. Reads originating from a top strand map to the C to T reference with read 1 reading in the forward direction and read 2 in the reverse, while bottom strand reads map to the G to A reference with read 2 mapping forward and read 1 in reverse. (H) The reads are binned by genomic locus and strand. Mutable positions and their conversions are recorded as a bit pattern. We then use those patterns to cluster the reads (see Figure 3).