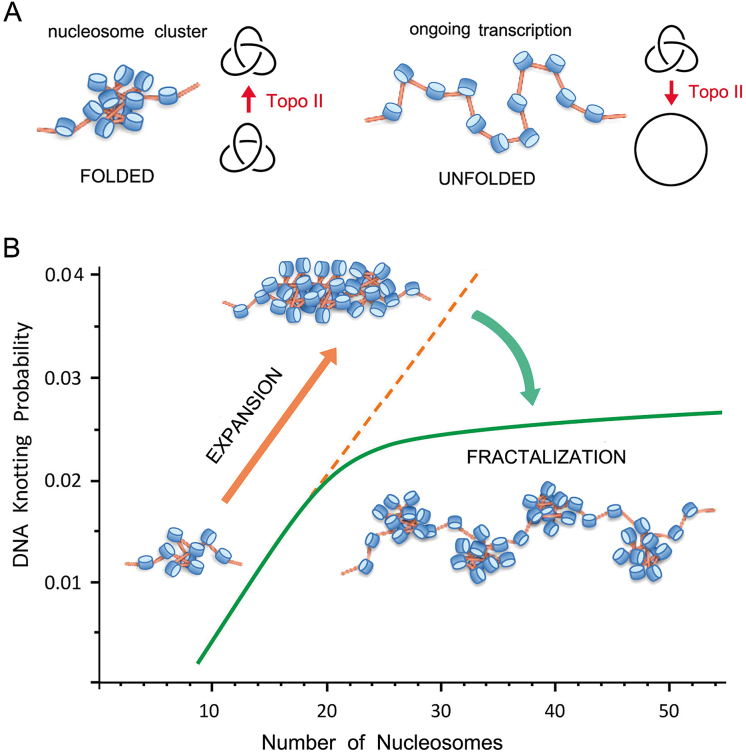
Figure 7.
Model of chromatin architecture inferred from DNA knotting probability. (A) Intricate folding of nucleosome arrays favors topo II-mediated knotting of intracellular DNA. Knotted fractions are reduced when nucleosomal clusters unfold during DNA transcription. (B) Uninterrupted expansion of nucleosomal fibers would produce proportional scaling of DNA knot formation (orange dashed line). Fractalization of the chromatin architecture minimizes instead the potentially harmful scaling of DNA entanglements (green line). The kPCHR data supports a fractal model, in which the ‘beads on a string’ architecture of the 10 nm nucleosomal fiber reiterates in its next level of organization by forming clusters of about 20 nucleosomes.