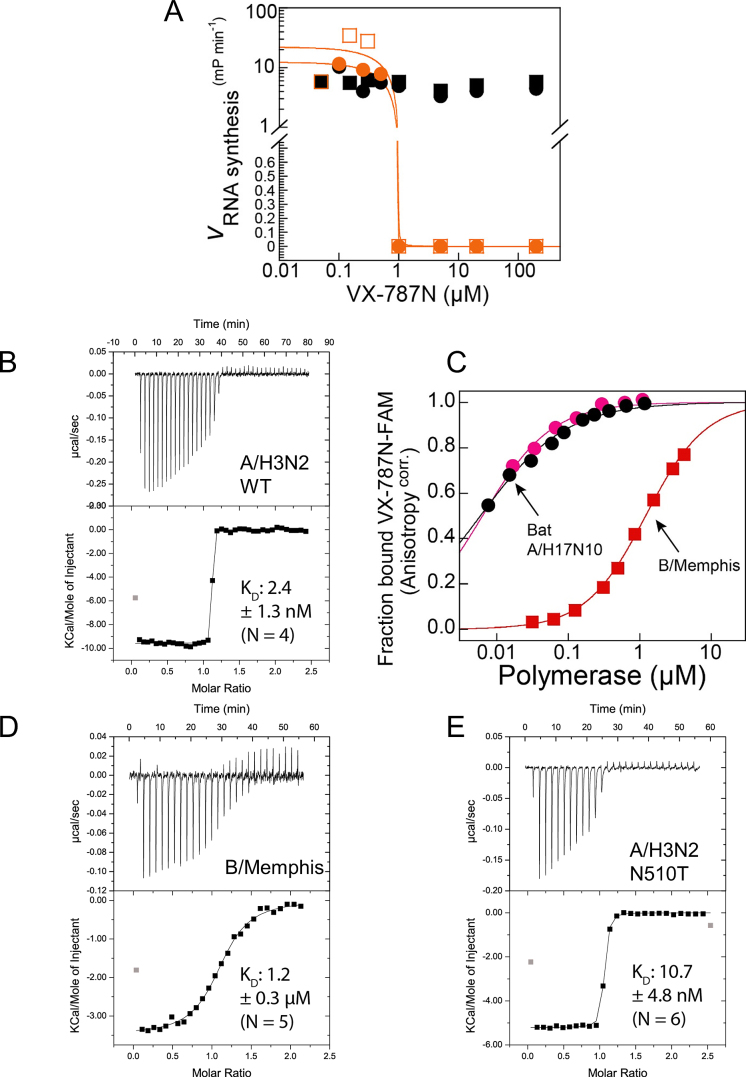
Figure 5.
Binding and inhibitory function of VX-787N or VX-787 to influenza A and B polymerase. (A) Effect of VX-787N on bat A/H17N10 influenza polymerase RNA synthesis activity. While VX-787N shows no effect on ApG primed RNA synthesis from the vRNA promoter (black), at an excess of VX-787N over polymerase and capped RNA primer, VX-787N completely inhibits capped RNA primed RNA synthesis from the vRNA promoter (orange) as expected. Squares and circles of identical color correspond to duplicate experiments. (B) Quantification of VX-787N-FAM binding to A/H5N1 influenza polymerase PB2 cap-binding domain or cap-midlink double domain. Interaction of A/H5N1 PB2 cap-binding domain (black) and cap-midlink double domain (red) with the fluorescently labelled VX-787N results in increased fluorescence polarization that was used to determine KDs of respectively 0.017 ± 0.004 μM and 0.007 ± 0.001 μM (error of the fitting routine of mean values of duplicate experiments; standard deviation of the mean is indicated). (C) Representative ITC data and curve fit to derive the affinity of VX-787 to A/H3N2 cap-midlink double domain. The derived KD is 2.4 ± 1.3 nM (N = 4). (D) Comparison of VX-787N-FAM binding to bat A/H17N10 polymerase or B/Memphis polymerase by fluorescence anisotropy assay giving KDs of respectively 0.0061 ±0.00021 μM (mean and standard deviation of duplicates; black and pink circles correspond to independent binding isotherms) and 1.2 ± 0.02 μM (representative binding isotherm ± error of the fitting routine; red squares). (E) Representative ITC data and curve fit to derive the affinity of VX-787 to B/Memphis cap-midlink double domain. The derived KD is 1.2 ± 0.3 μM (N = 5). (F) Representative ITC data and curve fit to derive the affinity of VX-787 to A/H3N2 (N501T) cap-midlink double domain. The derived KD is 10.7 ± 4.8 nM (N = 6).