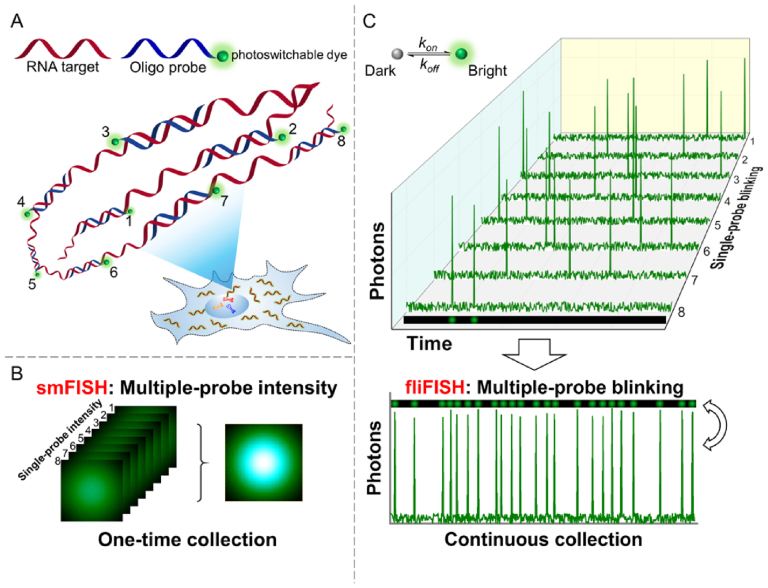
Figure 1.
Illustration of the fliFISH concept. (A) Multiple single-labeled probes (blue) are hybridized to a target RNA molecule (red), enabling the quantification and subcellular localization of the transcripts. (B) In conventional smFISH, integrated fluorescence intensity of multiple probes, acquired in a single image, is used to distinguish the target molecule from background noise. (C) In fliFISH, photoswitchable dye molecules attached to individual probes are stochastically turned on and off to generate photoblinking patterns as illustrated in the corresponding single-probe blinking traces. With each additional probe that is hybridized to the RNA molecule, the ensemble on-time fraction is increased in a predictable way. Knowing the number of probes that are designed to target the RNA molecule, the ensemble on-time fraction expected within a diffraction-limited area can be calculated and used as a threshold for a true signal.