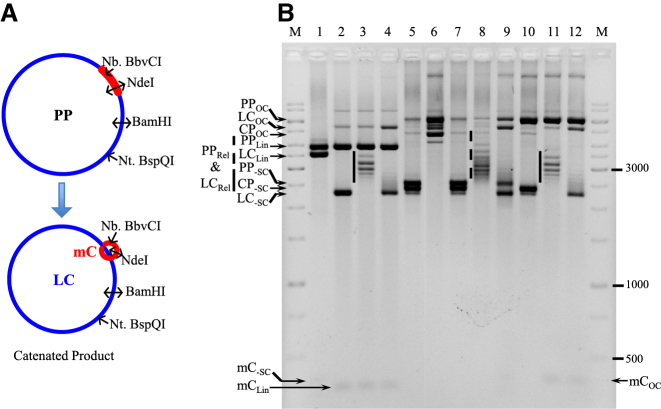
Figure 5.
Decatenation of multi-linked DNA circles by SsoTopA. (A) The PP used to produce catenated molecules (CP) is schematically represented. The catenated DNA is composed of the LC (in blue color) and the mC (in red color) which are interlinked. The different enzymes used to characterize the different forms of these DNA are indicated. (B) The multi-linked DNA substrate (lanes 5 and 7) produced from the parent plasmid pMC339 (PP) includes two major species, the PP itself and the catenane (CP), and one minor species, the decatenated LC and mC. The multi-linked DNA substrate was incubated for 1 h at 37°C with BamHI (lane 1), NdeI (lanes 2–4), Nt.BspQI (lane 6) or Nb.BbvCI (lanes 10–12). Sso TopA was added in lanes 3, 4, 8, 9, 11 and 12 and incubated for 20 min at 80°C (lanes 3, 8 and 11) or 90°C (lanes 4, 9 and 12) producing relaxed DNA. The vertical plain bars correspond to the large plasmid relaxed at 80°C by TopA and the vertical dashed line corresponds to a mix of large plasmid and catenane relaxed at 80°C by SsoTopA. The DNA was loaded onto a 1.7% agarose gel and ran at about 2.5 V cm−1 for 3 h. The different DNA bands were attributed according to their size and topological forms (OC, open circular; Lin, linear; Rel, relaxed; -SC, negatively supercoiled) as deduced from the control reactions. M corresponds to the GeneRuler 1 kb ladders (ThermoFisher).