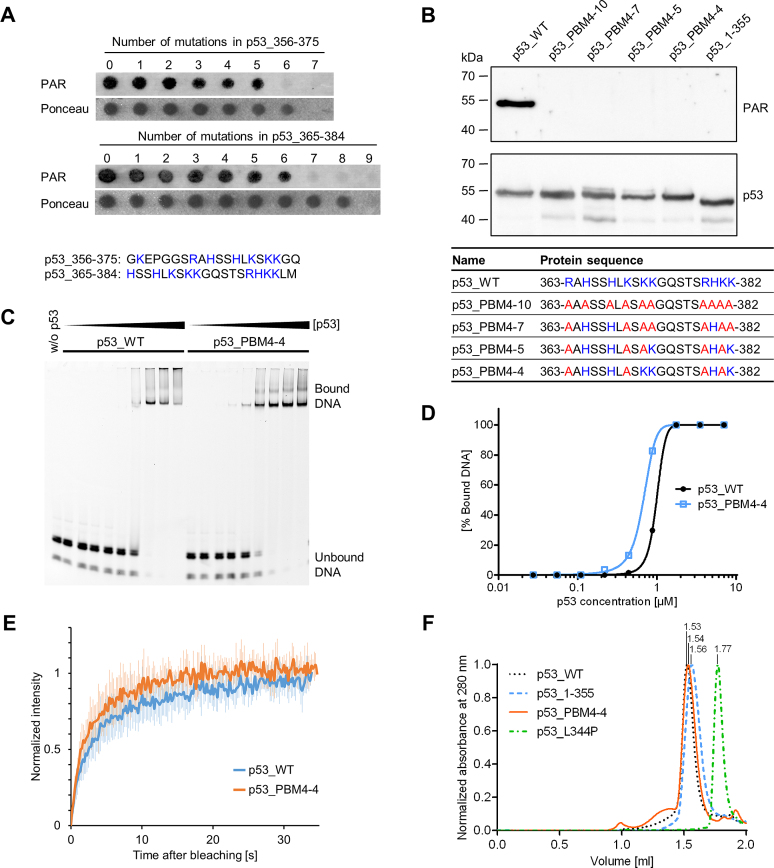
Figure 2.
Generation of a PAR-binding-deficient p53 mutant. (A) PAR overlay assay of PepSpot peptide array of CTD mutants with 1–9 aa exchanges. Ponceau S staining served as a loading control. Peptide size: 20 aa. Amino acids highlighted in blue were subjected to exchanges to alanines. The complete membrane with additional peptides and controls is shown in Supplementary Figure S2A and B. (B) PAR overlay assay after SDS-PAGE of full-length p53 with mutated PBM4. Exchanges of 10, 7, 5 or 4 aa were introduced into the CTD, which is shown in the table below. Basic aa (in blue) were subjected to exchanges to alanines (in red). (C) Electrophoretic mobility shift assay (EMSA) with p53_WT and p53_PBM4–4, binding to the fluorescently labeled DNA substrate REp21. (D) Densitometric quantification of (C). (E) FRAP analysis in H1299 cells reconstituted with GFP-tagged p53_WT or p53_PBM4–4. Recovery of GFP was measured after photobleaching. Means ± SD of six cells. (F) Analytical size-exclusion chromatography with p53_WT, p53_1–355, p53_PBM4–4 and a tetramerization-deficient p53 mutant (p53_L344P). Size-calculations are shown in Supplementary Figure S2E.