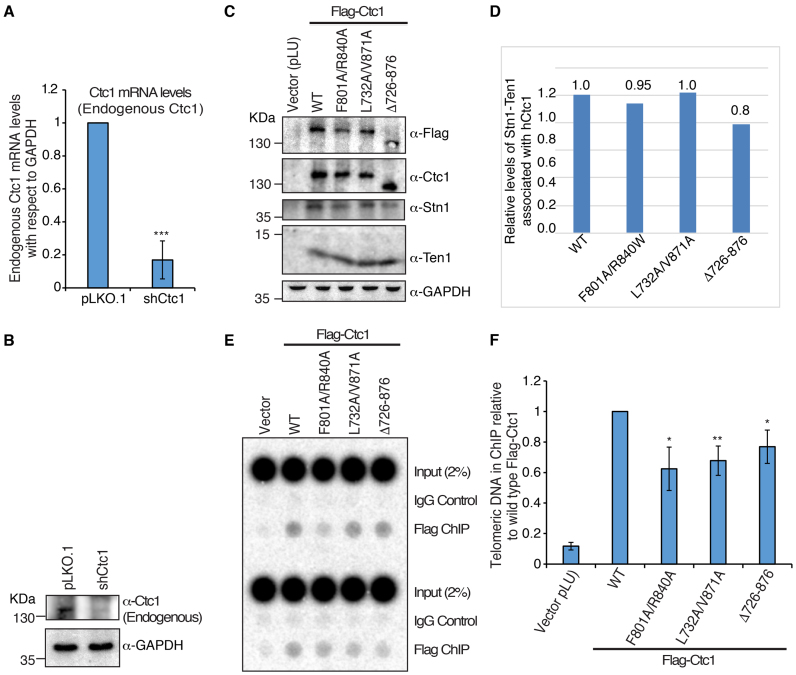
Figure 4.
Mutations in the hCtc1(OB) domain of hCtc1 affect its localization to telomeres. (A and B) qRT-PCR and western blot analysis of endogenous hCtc1 mRNA and protein levels, respectively, in 293T after shRNA knockdown. For qRT-PCR, results are the average of three independent experiments. Standard deviations are shown in the form of error bars; ***P < 0.0001 (calculated by two-tailed paired Student’s t-test). (C) Immunoprecipitation of hCtc1 stable cell lines established from 293T. Flag-tagged hCtc1 were isolated by flag beads and assayed by western blotting. Antibody for hCtc1 was used to detect the full-length hCtc1, hCtc1-(F801A/R840A), hCtc1-(L732A/V871A) and hCtc1-(Del726–876). GAPDH was used as the input control. (D) Bar graph showing the relative protein levels of Stn1–Ten1 complex pulled down by the full length, WT and mutant (double and deletion) hCtc1. Results are the average of three independent experiments. *P < 0.05 (calculated by two-tailed paired Student’s t-test). (E) Telomere localization of full length, WT and mutant (double and deletion) hCtc1 using crosslinking ChIP experiments in 293T cells expressing these proteins. ChIP DNA was assayed by dot blotting and hybridization with 32P-labeled probed specific for TTAGGG. (F) Bar graph showing the relative levels of full length, WT and mutant (double and deletion) hCtc1 proteins at telomeres. Results are the average of three independent experiments. *P < 0.05, **P < 0.001 (calculated by two-tailed paired Student’s t-test).