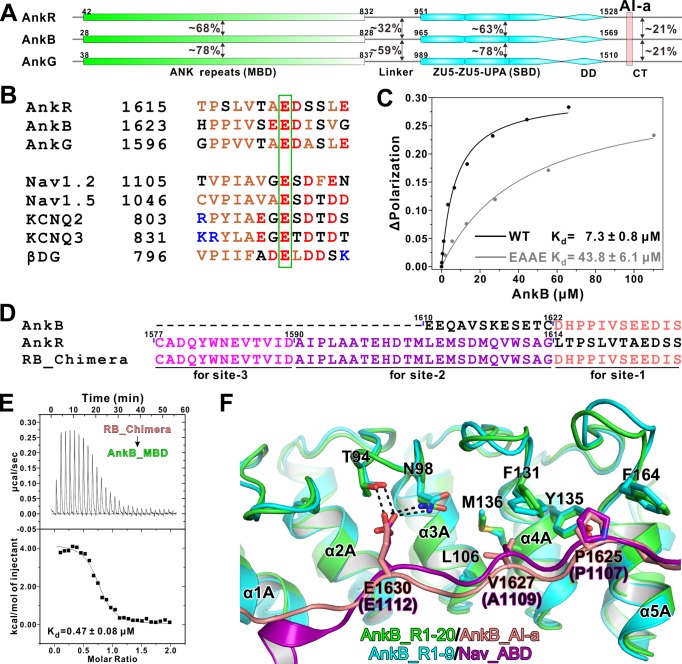
Figure 1. Biochemical and structural characterization of the AI-a/MBD interaction.
(A) Schematic diagram showing the domain organizations of AnkR/B/G and their amino acid sequence identities. (B) Amino acid sequence alignments of AI-a from AnkB/G with several MBD ‘site-1’ binding sequences from AnkR CT or other targets. The critical Glu residues are highlighted with a green box. (C) Fluorescence polarization-based measurement of the binding affinity between AnkB_AI-a and AnkB_MBD or its R1 charge reversal mutant (‘EAAE’). (D) The design of AnkR/B_Chimera construct for crystallization. The AnkB_AI-a sequence (colored in salmon) was fused to the C-terminus of ‘site-2, 3’ binding AnkR_CT (colored in purple and magenta respectively). (E) ITC result showing the strong interaction between AnkR/B_Chimera and MBD. (F) Structural comparison of the MBD ‘site-1’ bindings of AnkB_AI-a (colored in green and salmon) and Nav_ABD (colored in cyan and purple) showing that the two bindings essentially share the same mode. Residues critical for the interaction were highlighted with stick models. Hydrogen bonds were indicated with dashed lines (the same labeling method is used throughout the manuscript for all structural figures).