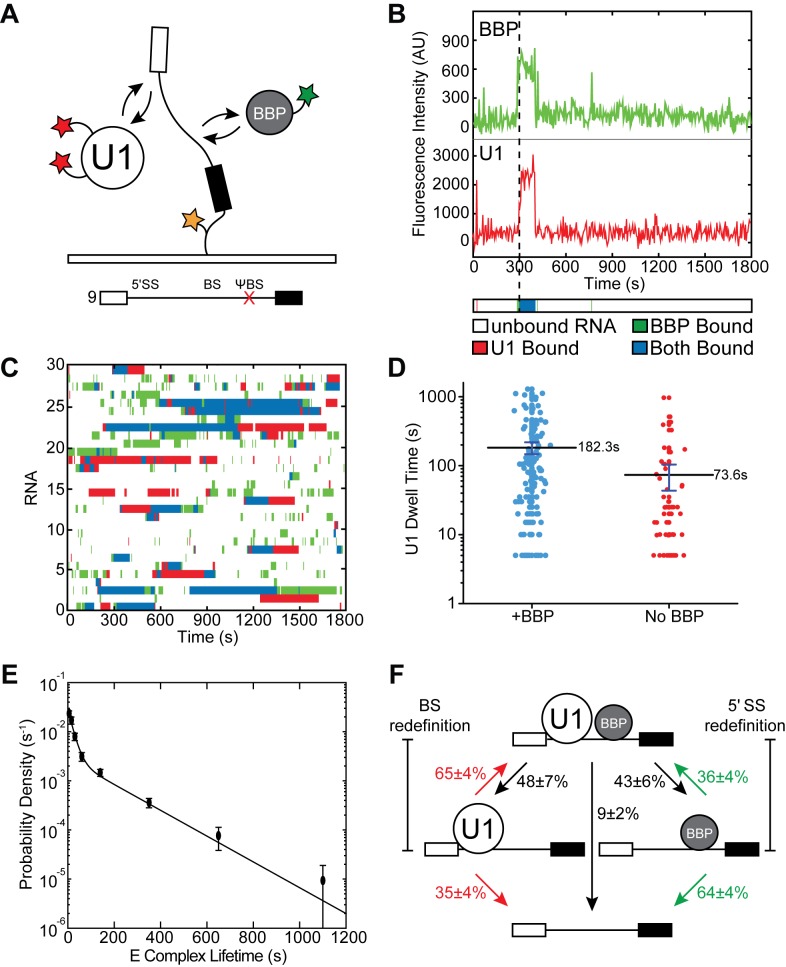
Figure 3. Three-color CoSMoS observation of U1 and BBP binding dynamics.
(A) Schematic of a three-color experiment in which U1 was labeled with two red-excited (Cy5) fluorophores, BBP was labeled with a single green-excited (Dy549) fluorophore, and the surface-tethered pre-mRNA was labeled with an Alexa488 fluorophore. Shown below is a graphic representation of the capped, immobilized RNA used in these three-color experiments. (B) Representative time records showing peaks in fluorescence intensity corresponding to colocalization of BBP (green) and U1 (red) with the same individual pre-mRNA molecule. The dashed line demarks the arrival of U1. Shown below the time record is the corresponding time ribbon in which U1 and BBP binding are represented by red and green bands, respectively. Colocalizations of U1 and BBP are represented by blue bands. (C) Rastergram depicting typical U1 and BBP binding events and colocalization on immobilized RNAs. (D) Measured U1 lifetimes in the presence or absence of BBP colocalization. The mean lifetimes are noted ±S.E. (E) Probability density histogram of dwell times for colocalized U1 and BBP complexes. The lines represent the fit of the distribution to an equation containing two exponential terms. Details of the fit parameters can be found in Supplementary file 5. (F) Routes for loss of either U1 or BBP fluorescent spots from immobilized RNAs following their colocalization. Percentages represent the fraction of U1/BBP complexes in which fluorescence disappeared by the indicated pathway (N = 173 initial colocalized U1/BBP complexes). Error bars in (E and F) represent the error in counting statistics as given by the variance of a binomial distribution.
Figure 3—figure supplement 1. Characterization of U1- and BBP-labeled yeast splicing extracts.
Figure 3—figure supplement 2. U1- and BBP-labeled WCE forms commitment complexes.
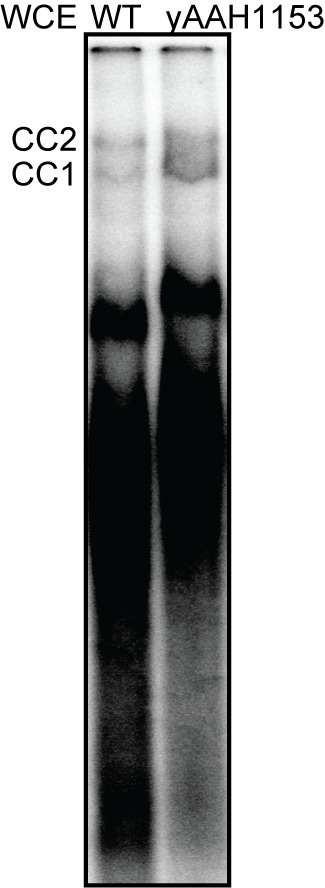
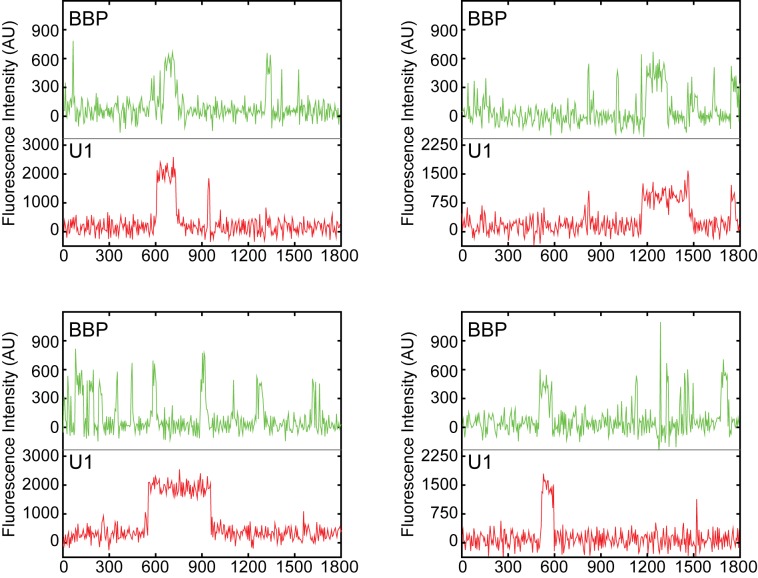
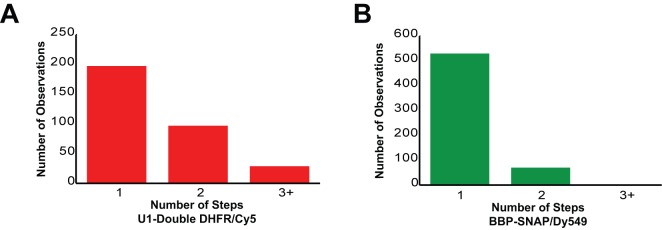
Figure 3—figure supplement 3. Examples of fluorescence intensity traces supplementing data shown in Figure 3B showing individual U1-DHFR subcomplexes and BBP-SNAPf co-localizing with surface-tethered RNAs.
Figure 3—figure supplement 4. Randomized control for analysis of U1 lifetimes during colocalization with BBP to complement Figure 3D.
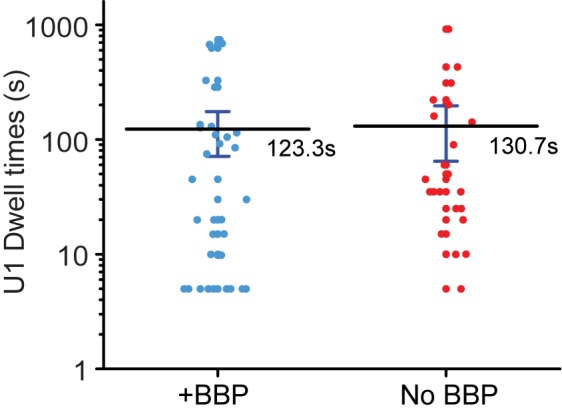
Figure 3—figure supplement 5. E complex lifetimes on RNAs containing or lacking the ΨBS.
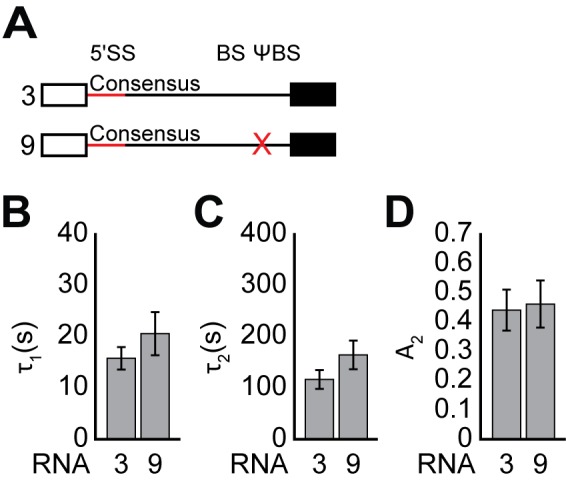
Figure 3—figure supplement 6. Pathways for E complex formation.