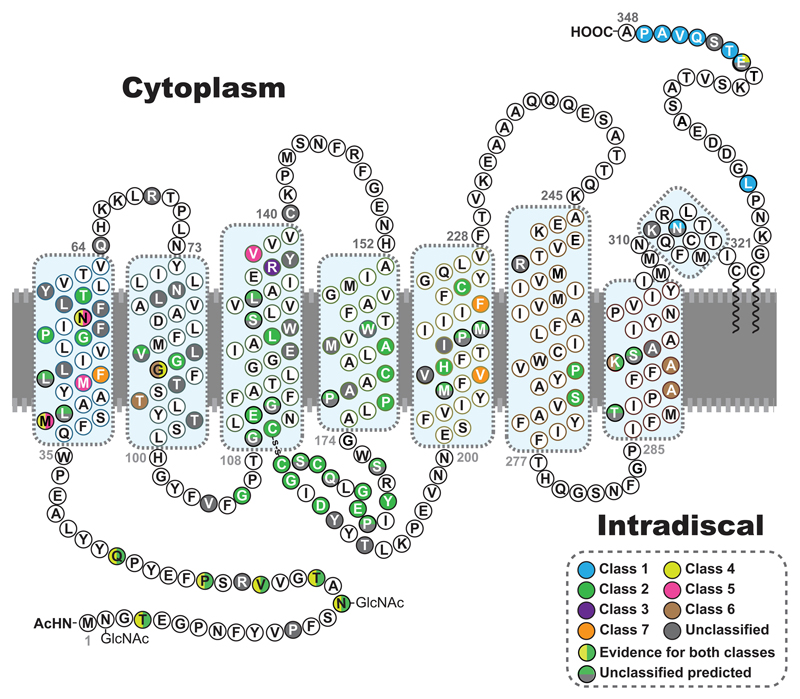
Figure 2. The position of rhodopsin mutations.
The primary amino acid sequence of human rhodopsin is shown as a secondary structure schematic to show the position of different classes of mutation. Class 1, post-Golgi trafficking and outer segment targeting (blue). Class 2 misfolding, ER retention and instability (green). Class 3, disrupted vesicular trafficking and endocytosis (violet). Class 4, altered post-translational modifications and reduced stability (yellow). Class 5 altered transducin activation (pink). Class 6 constitutive activation (brown). Class 7 dimerization deficiency (orange). Where there is evidence for more than one type of class it is shown with a vertical colour split. Uncategorised mutations, or those with no biochemical or cellular defects are shown in grey. Those with predicted effects on folding or binding from FoldX (Rakoczy et al., 2011) are shown with a horizontal colour split. The 8 alpha helix motifs of rhodopsin are highlighted by blue boxes.