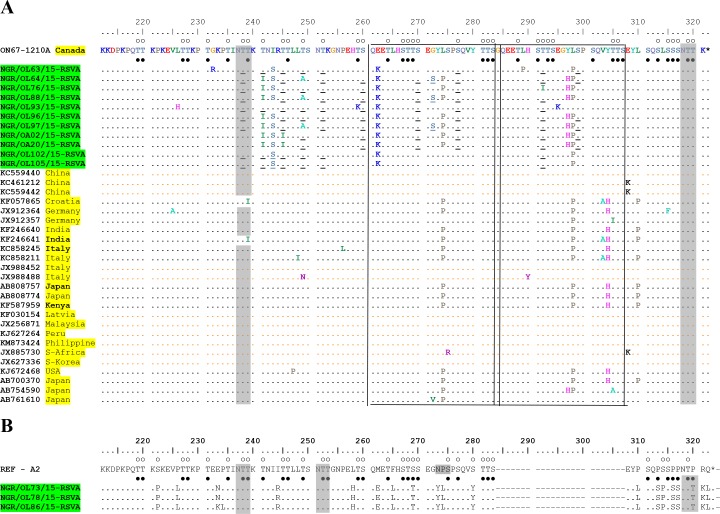
Fig 2. Alignment of deduced amino acid sequences of 2nd hypervariable region of HRSV-A strains.
A) Alignment of HRSV-A genotype ON1 sequences from different parts of the world. Alignments are shown and residues numbered relative to sequences of prototype ON1 strain ON67-1210A (GenBank accession no. JN257693). The strains from this study are highlighted. The country of isolation of other strains used in the alignment are also highlighted. Identical residues are indicated by dots and stop codons by asterisks. Potential N-glycosylation sites (N-X-T/S, where X is not a proline) are indicated by gray-shaded rectangles. Potential O-glycosylation sites of the prototype ON1 strain are indicated by unfilled circles, while black circles indicated the predicted O-glycosylation sites common in all Nigeria strains. Other predicted O- glycosylation sites, not found in all the strains in Nigeria are underlined. The two copies of the 23 amino acids duplicated sequences are framed by rectangles. B) Alignment of HRSV-A subtype NA2 from this study. Alignments are shown relative to the sequence of prototype strain A2 (GenBank accession number M11486). The identifier of strains from this study are highlighted. Residues are numbered relative to the amino acid sequences of prototype ON1 strain ON67-1210A (GenBank accession no. JN257693). Identical residues are indicated by dots, alignment gaps by dashes and stop codons by asterisks. Potential N-glycosylation sites (N-X-T/S, where X is not a proline) are indicated by shaded rectangles. Potential O-glycosylation sites of the prototype A2 strain were indicated by unfilled circles, while black circles indicated the predicted O-glycosylation sites in the Nigeria NA2 strains.