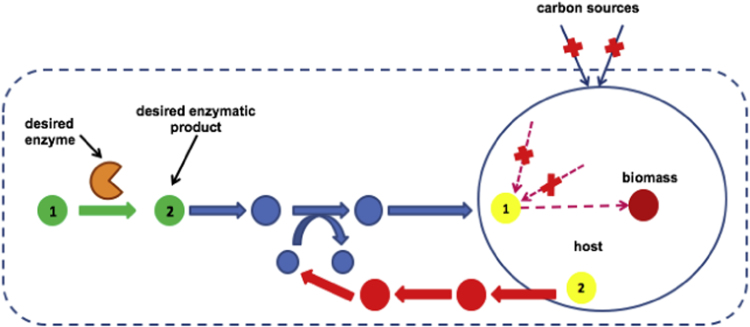
Fig. 2.
Illustration of SelFi implementation. The large round circle indicates boundaries of the wild-type E. coli, and the dotted box indicates boundaries of the cell after co-expression of the selection system. The desired enzyme catalyzes a reactant (green number 1) to a desired product (green number 2). A consumption pathway (blue) from the desired product to a metabolite (yellow number 1) within the wild-type E. coli is constructed using retroProPath to consume the desired product. A supporting pathway (orange) from a native metabolite (yellow number 2) in the wild type to a cofactor on the reactant side of a consumption pathway is constructed using ProPath, if needed. An “x” within the cell indicates a knockout, and an “x” outside the cell indicates eliminating carbon sources. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article).