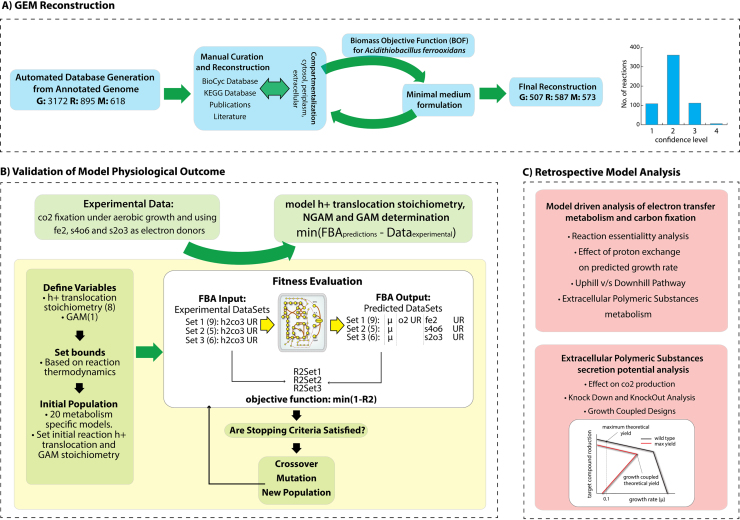
Fig. 1.
General workflow to generate, validate and further analysis of iMC507: (A) the workflow detailing the iterative model building procedure used to generate iMC507. The reconstruction process was initiated based on the annotated genome generated by PathoLogic. Manual curation and reconstruction was performed by using the Insilico Biotechnology software aided by the KEGG and MetaCyc databases. Publications and literature sources were used to refine the network content, assigning a specific confidence score to each reaction. The reconstructed network in conjunction with the BOF, were used to formulate a minimum medium for the three different metabolism studied. (B) Proton translocation for 8 different membrane reaction and GAM stoichiometry were estimated using a genetic algorithm. Experimental data for growth under three different electron donors and FBA was used to decipher the model parameters that best represent cell behavior. Based on these results the iMC507 network was validated with physiological data for growth under ferrous ion, tetrathionate and thiosulfate. (C) To study the most relevant aspects of the electron transfer metabolism and carbon fixation, a retrospective model analysis was performed by using iMC507. Further analysis on EPS secretion potential for growth-coupled production through knock-outs was performed.