Figure 2.
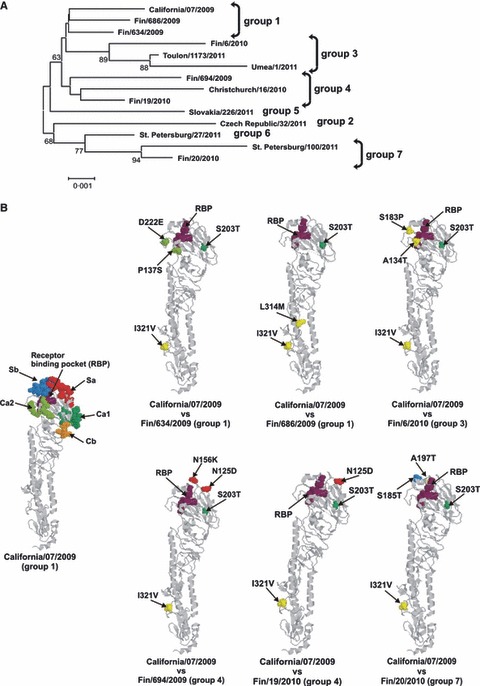
(A) Influenza A(H1N1)pdm09 strains isolated in Finland cluster in the same genetic groups with WHO reference strains in the phylogenetic tree of the HA. All sequences included in the phylogenetic tree constitute the entire 1698 nucleotide long coding region of HA. The horizontal lines are proportional to the number of nucleotide changes. (B) Schematic representation of amino acid differences in the HA molecule between the Finnish influenza A(H1N1)pdm09 viruses and the vaccine virus, A/California/07/2009. On the left, a side view of the monomeric structure of HA molecule of influenza A(H1N1(2009) (A/California/04/2009; RCSB Protein Bank accession number 3LZG) with previously identified H1 protein‐specific antigenic sites (Sa in red, Sb in blue, Ca1 in darker green, Ca2 in lighter green, and Cb in orange) of influenza A(H1N1) viruses and with the receptor binding pocket (RBP, purple) is shown. The amino acid changes of Finnish A(H1N1)pdm09 viruses compared with A/California/07/2009, the vaccine strain, are illustrated in the monomeric HA structure and colored as in A/California/07/2009 structure. Amino acid changes outside the antigenic sites are shown in yellow. Changes are illustrated by amino acid residue number and with serial number of virus where the respective amino acid change has been observed.
