Abstract
CDGSH iron sulfur domain 2 (CISD2) has been found to be important in carcinogenesis. However, the role of CISD2 in glioma remains to be elucidated. The present study aimed to investigate the role of CISD2 in glioma using the reverse transcription-quantitative polymerase chain reaction, western blotting, co-immunoprecipitation assay, immunofluorescence staining and other methods. The results demonstrated that the mRNA and protein levels of CISD2 were found to be upregulated in glioma tissues, compared with the levels in matched normal tissues. Clinical data analysis showed that the level of CISD2 was negatively correlated with the survival rates of patients with glioma. In addition, high levels of CISD2 were associated with advanced clinical stage, relapse, vascular invasion and increased tumor size. The inhibition of CISD2 suppressed the proliferation and survival of glioma cells in vitro and in vivo. Mechanistically, it was found that small interfering RNA-induced knock down of CISD2 inhibited the proliferation of glioma cells through activating beclin-1-mediated autophagy. The results also revealed that CISD2 was a target of microRNA (miR)-449a. Together, the results of the present study demonstrated that CISD2 was increased in glioma samples and was associated with poor prognosis and aggressive tumor behavior. The miR-449a/CISD2/beclin-1-mediated autophagy regulatory network contributed to the proliferation of glioma cells. Targeting this pathway may be a promising strategy for glioma therapy.
Keywords: glioma, CDGSH iron sulfur domain 2, autophagy, beclin-1, microRNA-449a
Introduction
Glioma is one of the most common malignant tumors threatening human health and has increased in incidence (1,2). Even with aggressive treatments, the average 5-year survival rate remains poor (3,4). Detailed investigations have been performed to understand the pathogenesis of glioma. However, the underlying molecular network involved in the initiation and progression of glioma remains to be fully elucidated.
CDGSH iron sulfur domain 2 (CISD2) belongs to the CDGSH iron sulfur domain protein family (5). CISD2 contains a transmembrane domain, a CDGSH domain and a conserved amino acid sequence for iron binding. Located in the outer membrane of mitochondria, CISD2 is important for mitochondrial integrity and lifespan (6–8). CISD2 deficiency leads to mitochondrial damage, following which autophagy is induced to eliminate the impaired mitochondria (9). Compared with young mice, the levels of CISD2 are significantly lower in older mice. In addition, CISD2-knockout mice exhibit a significant premature aging phenotype, characterized by opaque eyes, blindness, lordokyphosis, osteopenia and skin atrophy (10). CISD2 has been demonstrated to be important in tumor cells. CISD2 is reported to be elevated in human epithelial breast cancer cells, and significantly promotes cell proliferation and tumor growth (11). CISD2 has also been identified as a novel marker correlating with metastasis and prognosis in patients with early-stage cervical cancer (12). However, the role of CISD2 in glioma remains to be elucidated.
Autophagy is a conserved process, which is responsible for the turnover of long-life proteins or for the removal of damaged organelles in eukaryotic cells (13). Autophagy is typically activated under conditions of starvation, and autophagy marker proteins include beclin-1, light chain 3 (LC3) and p62 (14–16). p62 is a selective substrate of autophagy, and its accumulation is observed when autophagy is inhibited (17). The role of autophagy in cancer differs depending on the situation (18). Studies have shown that the inhibition of autophagy promotes cancer initiation, however, others have shown that it can also suppress the growth of certain malignancies, including breast cancer and hepatocellular carcinoma (19–22). Therefore, it is necessary to investigate the involvement of autophagy in the function of CISD2 in glioma.
In the present study, the levels of CISD2 in glioma tissues were evaluated, and the association between levels of CISD2 and the prognosis of patients with glioma was examined. In addition, the role of CISD2 in the proliferation and carcinogenesis of glioma cells was investigated in vitro and in vivo. The downstream signaling pathway underlying the oncogenic role of CISD2 in glioma cells was identified and the results provided evidence that CISD2 was a direct target of miR-449a. Taken together, the data suggested that CISD2 is important in the proliferation of glioma and indicated that CISD2 may be a novel therapeutic target for the treatment of glioma.
Materials and methods
Clinical sample collection
The present study was reviewed and approved by the ethical review board of Xinxiang Medical University (Xinxiang, China). All applicable international, national, and/or institutional guidelines for the care and use of animals were followed. All procedures performed involving human participants were in accordance with the ethical standards of the institutional and national research committee, and with the 1964 Helsinki Declaration and its later amendments or comparable ethical standards. The study was performed following the provision of written informed consent from patients. A total of 72 fresh glioma tissues and their adjacent non-glioma tissues were collected during surgery between December 2014 and December, 2015. The levels of CISD2 were evaluated in these paired tissues. In order to determine the association between CISD2 and the prognosis of patients with glioma, 120 paraffin-fixed glioma specimens were collected from patients between January 2008 and January 2010, and these patients were followed up 40 months later.
Cell culture
The U87 glioma cell line was purchased from the Chinese Academy of Sciences (Shanghai, China) and was cultured in DMEM (Thermo Fisher Scientific, Inc., Waltham, MA, USA) supplemented with 10% FBS (Thermo Fisher Scientific, Inc.). Please note that the U87 cell line is known to be cross-contaminated with another cell line, which is most likely to be a glioblastoma cell line (23). The cells were maintained in a humidified 37°C incubator containing 5% CO2. For autophagy inhibition, the cells were treated with 1 mM 3-MA (Sigma-Aldrich; Merck KGaA, Darmstadt, Germany) for 24 h at 37°C.
Tumor graft
BALB/c nude male mice (6 weeks old) were purchased from the Laboratory Animal Center of Xinxiang Medical University. All mice were housed in a strictly pathogen-free conditions at room temperature with free access to food and water and 12 h/12 h light/dark cycle. The protocols for the experiments involving mice complied with the Guide for the Care and Use of Laboratory Animals (National Institutes of Health, Bethesda, MD, USA) and approved by the Animal Ethics Committee of the Xinxiang Medical University.
Following being transfected with the small interfering RNA to knock down CISD (si-CISD) or a scramble construct, the U87 cells (5×107) were harvested and subcutaneously inoculated into the right groin of the nude mice (n=3 per group). Following growth for 40 days, the formed tumors were carefully excised. The weight of the formed tumor was measured and the volume was calculated using the following formula: Length × width2 × π/6.
Reverse transcription-quantitative polymerase chain reaction (RT-qPCR) analysis
TRIzol reagent (Thermo Fisher Scientific, Inc.) was used to extract RNA from samples. Equal quantities of RNA were then reverse transcribed into cDNA with SuperScript® IV Reverse Transcriptase (Thermo Fisher Scientific, Inc.). A total of 2 µg CISD2 cDNA was amplified under the following thermal conditions: 95°C for 10 min; followed by 40 cycles of 95°C for 15 sec and 60°a for 1 min; and 4°C holding; the total duration was 1 h 48 min. GAPDH was used as an internal control. The final results were calculated using the 2−ΔΔCq (24) method and presented as fold changes. The following primers were used: CISD2, forward 5′-GCAAGGTAGCCAAGAAGTGC-3′ and reverse 5′-CCCAGTCCCTGAAAGCATTA-3′; GAPDH, forward 5′-GCGAGATCGCACTCATCATCT-3′ and reverse 5′-TCAGTGGTGGACCTGACC-3.
Western blot analysis
Briefly, the tissues were lysed with radioimmunoprecipitation assay lysis buffer containing inhibitor cocktail for protease and phosphatase (Thermo Fisher Scientific, Inc.). The concentration of protein sample was determined using the bicinchoninic acid method. Following boiling for 15 min, 15-µg protein samples were separated on a 10% SDS gel, blocked with 5% nonfat milk and transferred onto a PVDF membrane (EMD Millipore, Billerica, MA, USA). The membrane was incubated with primary antibody overnight at 4°C. Following washing with TBST five times, the membrane was incubated with secondary antibody for 2 h at 27°C. The membrane was washed with TBST five times, following which the membrane was visualized with Pierce™ Enhanced Chemiluminescence Plus Western Blotting Substrate (Thermo Fisher Scientific, Inc.). GAPDH was used as an internal control. The protein band intensity was measured using Image J software (version 1.49; National Institutes of Health, Bethesda, MD, USA).
The primary antibodies used in the present study were all purchased from Sigma-Aldrich (Merck KGaA) and were as follows: Anti-CISD2 (1:1,000; cat no. AV44552), anti-LC3-II (1:800; cat. no. ABC432), anti-beclin-1 (1:1,000; cat. no. SAB1306484), anti-autophagy related 7 (Atg7) (1:1,000; cat. no. MABN1124), anti-p62 (1:1,000; cat. no. MABC32) and anti-GAPDH (1:1,000; cat. no. G9545). The secondary antibodies used were as follows: Horseradish peroxidase (HRP)-labeled goat anti-rabbit immunoglobulin (Ig)G (1:3,000; cat. no. A0208; Beyotime Institute of Biotechnology, Haimen, China) and HRP-labeled goat anti-mouse IgG (1:3,000; cat. no. A0216; Beyotime Institute of Biotechnology).
Immunohistochemistry
Immunohistochemical staining was performed on the collected clinical glioma tissues sections to detect the levels of CISD2. Briefly, formalin and paraffin were used to fix and embed the tissues, respectively. The samples were heated to retrieve the antigen. The sections were incubated with anti-CISD2 primary antibody (1:500; cat. no. AV44552; Sigma-Aldrich; Merck KGaA) at 4°C overnight. Following three washes with PBS, the sections with incubated with HRP-labeled goat anti-rabbit IgG (1:1,000; cat. no. A0516; Beyotime Institute of Biotechnology) for 2 h. Following three washes with PBS, the sections were visualized with DBA solution, followed by counterstaining of nuclei with hematoxylin. Images were captured using a microscope (Eclipse Ci-E; Nikon, Tokyo, Japan).
Co-immunoprecipitation assay
A Pierce Co-IP kit (Thermo Fisher Scientific, Inc.) was used to examine the binding activity between CISD2 and beclin-1 in the indicated groups. The general procedure was performed according to the manufacturer's protocol, as previously described (25). The protein extracts were precipitated using anti-CISD2 (Sigma-Aldrich; Merck KGaA), and the precipitated protein was evaluated using western blot analysis with anti-beclin-1 (Sigma-Aldrich; Merck KGaA).
Immunofluorescence
Immunofluorescence was also used to determine the levels of CISD2 in the glioma tissue sections. Briefly, following fixation with 4% paraformaldehyde, the sections were incubated with anti-CISD2 primary antibody overnight at 4°C. Following three washes with PBS, the sections were incubated with Cy3-labeled goat anti-rabbit IgG (1:1,000; cat. no. A0516; Beyotime Institute of Biotechnology) for 2 h at 27°C. Images were captured using a laser confocal microscope (A1; Nikon Corporation, Tokyo, Japan).
Plasmid transfection
Briefly, when the cells reached a confluence of 70%, the cells were transfected with negative scramble siRNA, a CISD2-overexpression plasmid or an si-CISD2 and/or si-beclin-1 plasmid using Lipofectamine® 2000 transfection reagent (Thermo Fisher Scientific, Inc.). After 6 h, the DMEM was replaced with normal medium. All plasmids were purchased from GenePharma Co., Ltd. (Shanghai, China).
Detection of proliferation rates
The U87 glioma cells were transfected with the scramble or si-CISD2 plasmid and seeded into a 96-well plate at a confluence of 30% (3×104 cells/well). At the indicated time points, the number of cells in each well was determined using a Scepter Handheld Automated Cell Counter (EMD Millipore).
TUNEL assay
A TUNEL assay was used to measure apoptosis. Briefly, cells grown on a cover slip were fixed with 4% paraformaldehyde. Following washing with PBS, the cells were incubated with 0.3% H2O2 to block endogenous peroxidase activity. The cells were then incubated with TUNEL reaction solution (Sigma-Aldrich; Merck KGaA) for 1 h at 37°C. Images were captured under a laser confocal microscope (A1; Nikon).
Wound-healing assay
The U87 glioma cells were transfected with scramble or si-CISD2 plasmid and seeded into a 6-well plate at a confluence of 30% (3×104 cells/well). When cell confluence reached 95%, the cells were starved for 12 h. A 100-ml pipette tip was then used to scratch a straight line in the cell layer. Following incubation for another 24 h, the cells were fixed and images were captured under a microscope (Eclipse Ci-E; Nikon Corporation). The length of the wound was measured using Image J software.
Luciferase reporter assay
CISD2 wild-type 3-untranslated region (UTR) and mutated 3′-UTR constructs were sub-cloned into the pGL3 Luciferase Promote Vector (Sangon Biotech Co., Ltd., Shanghai, China) with XbaI and NotI restriction sites. Using Lipofectamine® 2000 transfection reagent (Thermo Fisher Scientific, Inc.), the pGL3 vector containing the CISD2 wild-type 3′-UTR or mutated form was co-transfected with or without miR-449a mimic (GenePharma Co., Ltd.) into the U87 cells. At 48 h post-transfection, the luciferase activity was measured using a Luciferase Reporter Assay kit (Sangon Biotech Co., Ltd.).
Statistical analysis
Data are expressed as the mean ± standard deviation of at least three independent experiments. Comparisons between two groups were analyzed using Student's t-test (two-tailed). Comparisons among groups were analyzed using one-way analysis of variance followed by Student-Newman-Keuls test. All analyses were performed with SPSS 19.0 software (IBM SPSS, Armonk, NY, USA). P<0.05 was considered to indicate a statistically significant difference.
Results
CISD2 is increased in glioma samples and negatively correlated with survival rates of patients
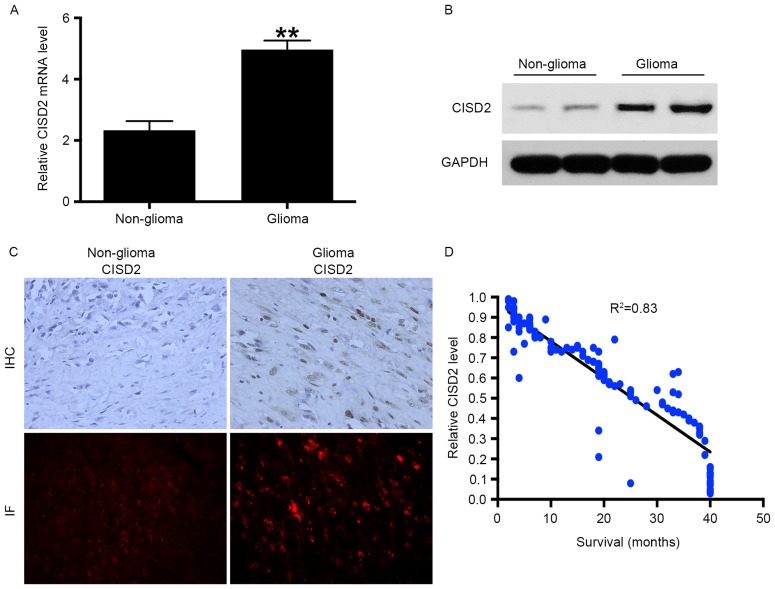
RT-qPCR analysis was performed to measure the mRNA levels of CISD2 in 72 fresh glioma tissue samples and corresponding non-glioma tissue samples. The results showed that the mRNA level of CISD2 was significantly increased in the glioma tissues, compared with that in the non-glioma tissues (Fig. 1A). In addition, elevated expression of CISD2 in glioma tissues at the protein level was confirmed using western blot analysis (Fig. 1B), immunohistochemistry and immunofluorescence (Fig. 1C). Taken together, the above data demonstrated that the mRNA and protein levels of CISD2 were markedly elevated in glioma tissues, compared with non-glioma tissues. As CISD2 was significantly increased in the glioma tissues, it was hypothesized that CISD2 may be a predictor of the survival rates of patients with glioma. A total of 120 paraffin-fixed glioma specimens were collected between January 2008 and January 2010 and the corresponding survival rates of the patients were recorded. The results showed that the level of CISD2 was negatively correlated with the survival rates of the patients (Fig. 1D). The associations between the level of CISD2 and clinicopathological characteristics were also analyzed in these patients (Table I). A high level of CISD2 was associated with advanced clinical stage (P<0.05), relapse (P<0.05), vascular invasion (P<0.05) and increased tumor size (P<0.05), but not differentiation (P>0.05).
Figure 1.
CISD2 is increased in glioma samples and negatively correlated with patient survival rate. (A) 72 fresh glioma tissues and their corresponding non-glioma tissues were collected, and mRNA levels of CISD2 were determined using reverse transcription-quantitative polymerase chain reaction analysis. (B) Protein levels of CISD2 in glioma and non-glioma tissues were determined using western blot analysis. GAPDH was used as an internal control. (C) Levels of CISD2 were also evaluated using IHC and IF (Red=CISD2) (magnification, ×200). (D) A total of 120 paraffin-fixed glioma specimens were analyzed and the survival rates of the corresponding patients were recorded. The results showed that the level of CISD2 was negatively correlated with the survival rates of patients with glioma. Data are presented as the mean ± standard deviation from at least three independent experiments. **P<0.01, compared with the control group. CISD2, CDGSH iron sulfur domain 2; IHC, immunohistochemistry; IF, immunofluorescence.
Table I.
Association between the expression of CISD2 and clinicopathological parameters in 120 patients with glioma.
| Expression of CISD2 | ||||
|---|---|---|---|---|
| Parameter | Total (n) | High (n) | Low (n) | P-value |
| Total | 120 | 83 | 37 | |
| Age (years) | NS | |||
| <50 | 53 | 36 | 17 | |
| <50 | 67 | 47 | 20 | |
| Gender | NS | |||
| Male | 56 | 38 | 18 | |
| Female | 64 | 45 | 19 | |
| Clinical stage | <0.05 | |||
| I–II | 33 | 11 | 22 | |
| III–IV | 87 | 72 | 15 | |
| Relapse | <0.05 | |||
| No | 41 | 20 | 21 | |
| Yes | 79 | 63 | 16 | |
| Vascular invasion | <0.05 | |||
| No | 72 | 40 | 32 | |
| Yes | 48 | 43 | 5 | |
| Differentiation | >0.05 | |||
| Well | 77 | 53 | 24 | |
| Moderate | 43 | 29 | 14 | |
| Tumor size | <0.05 | |||
| <3 cm | 62 | 36 | 26 | |
| >3 cm | 58 | 47 | 11 | |
CISD2, CDGSH iron sulfur domain 2; NS, not significant.
CISD2 promotes the proliferation and survival of glioma cells
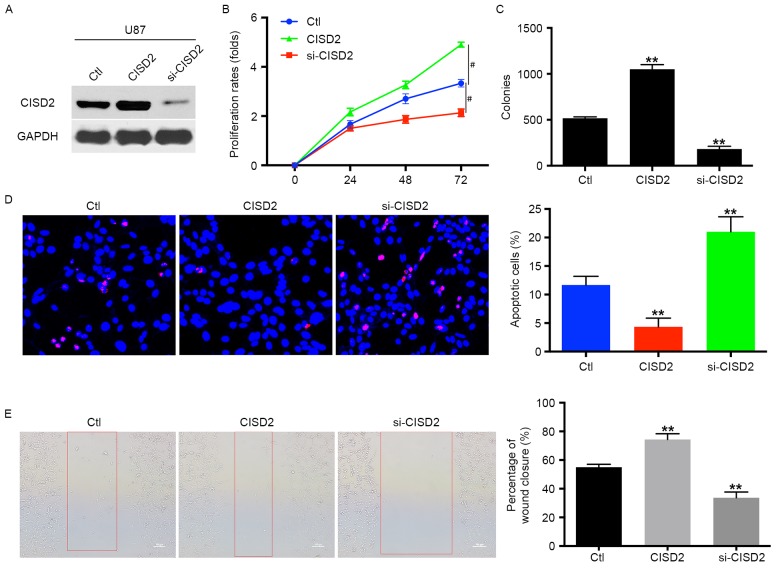
To examine the role of CISD2 in glioma cells, the level of CISD2 was upregulated using a CISD2 expression construct or knocked down using siRNA in U87 glioma cells. These effects were validated using western blot analysis (Fig. 2A). Compared with the vector control cells, the upregulation of CISD2 significantly increased the proliferation rate of the U87 cells (Fig. 2B). The overexpression of CISD2 also significantly increased the mean number of colonies in the colony formation assay (Fig. 2C). However, the knock down of CISD2 significantly reduced the proliferation rate and number of colonies of glioma cells, as indicated in the proliferation and colony formation assays, compared with the control or CISD2 upregulation groups (Fig. 2B and C). In addition, the TUNEL assay revealed that the inhibition of CISD2 markedly increased the apoptosis of U87 cells, compared with the control or CISD2 overexpression groups (Fig. 2D). The wound-healing assay showed that, compared with the control group, the upregulation of CISD2 enhanced the invasive ability of the U87 cells, whereas si-CISD2 significantly inhibited the invasion of U87 cells (Fig. 2E). Taken together, these results suggested that CISD2 promoted the proliferation and survival of glioma cells.
Figure 2.
CISD2 promotes proliferation and survival of glioma cells. (A) In U87 cells, a CISD2 expression construct was applied to upregulate CISD2, whereas siRNA was used to knock down CISD2 (si-CISD2). The efficiency of modulating CISD2 was confirmed using western blot analysis. The effects of modulating CISD2 on (B) proliferation rate and (C) colony formation were evaluated. (D) A TUNEL assay was used to detect the apoptosis in U87 cells. Compared with the control group, CISD2-overexpression decreased apoptosis, whereas si-CISD2 increased apoptosis. (the magnification is 200 times). (E) A wound healing assay showed that the overexpression of CISD2 enhanced the invasive ability of U87 cells, whereas si-CISD2 weakened invasive ability, compared with the control group (magnification, ×100). Data are presented as the mean ± standard deviation from at least three independent experiments. **P<0.01, compared with the Ctl group; #P<0.05, compared with the indicated groups. CISD2, CDGSH iron sulfur domain 2; si-CISD2, small interfering RNA targeting CISD2; Ctl, control.
CISD2 silencing inhibits carcinogenesis of glioma cells in a xenograft model
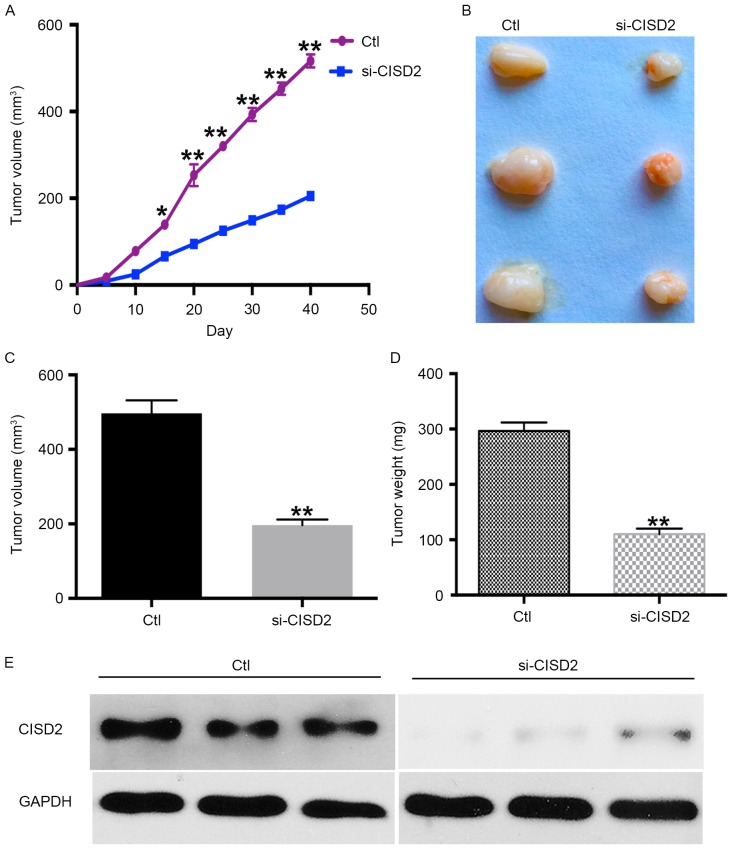
The present study also established a xenograft model in BALB/C nude mice using U87 cells to determine the in vivo effects of silencing CISD2. The results showed the silencing of CISD2 led to a significantly slower growth rate, compared with that in the control group (Fig. 3A). In addition, the CISD2-deficit U87 cells formed smaller tumors, compared with those in the control vector-transfected cells (Fig. 3B). The average volume and weight of tumors were significantly lower in the CISD2-deficit group, compared with those in the control group (Fig. 3C and D). Western blot analysis confirmed that CISD2 was effectively knocked down in the formed tumors of the si-CISD2 group, compared with that in the control group (Fig. 3E). Taken together, these data demonstrated that silencing of CISD2 significantly reduced the ability of glioma cells to form tumors in vivo.
Figure 3.
CISD2 silencing inhibits carcinogenesis of glioma cells in a xenograft model. Following transfection with the si-CISD or scramble construct, U87 cells were harvested and subcutaneously inoculated into the right groin of nude mice (n=3/group). (A) Tumor growth curve. (B) Following growth for 40 days, the formed tumors were carefully excised. (C) Weights and (D) volumes of the formed tumors were determined. (E) Levels of CISD2 in the formed tumors were determined using western blot analysis. Data are presented as the mean ± standard deviation from at least three independent experiments. *P<0.05 and **P<0.01, compared with the Ctl group. CISD2, CDGSH iron sulfur domain 2; si-CISD2, small interfering RNA targeting CISD2; Ctl, control.
CISD2 silencing activates autophagy in glioma cells
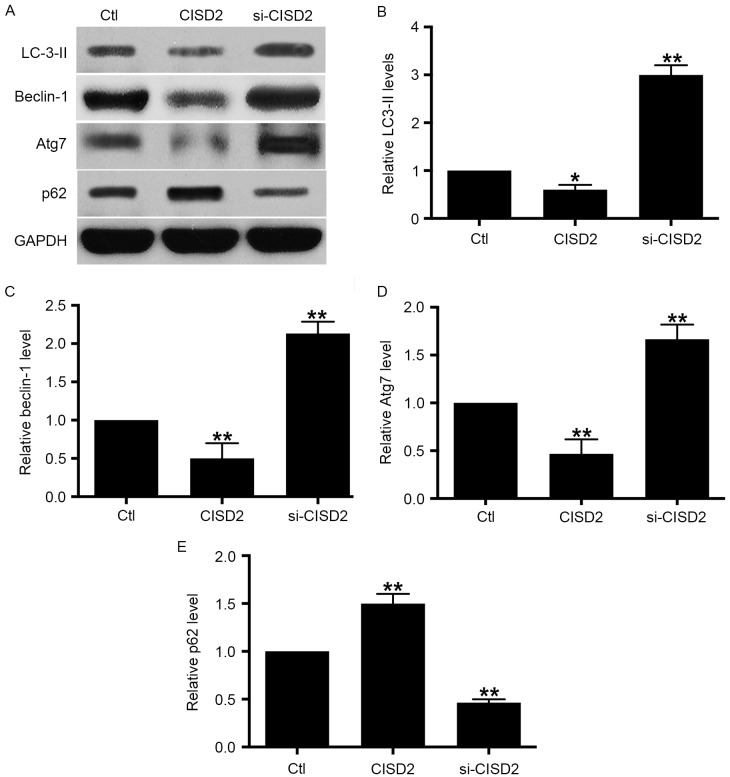
In order to examine the involvement of autophagy in the tumor-inhibitory effects of si-CISD2 in glioma cells, CISD2 was manipulated and the levels of autophagy activity markers were evaluated using western blot analysis. The resulting data showed that, compared with the control group, silencing of CISD2 led to significant increases in the levels of LC3-II, beclin-1 and Atg7, and a decrease in selective autophagy target p62; this was reversed by the overexpression of CISD2 (Fig. 4A-E). The above data demonstrated that the silencing of CISD2 activated autophagy whereas the overexpression of CISD2 inhibited autophagy in glioma cells.
Figure 4.
Silencing CISD2 activates autophagy in glioma cells. (A) In the U87 cells, the CISD2 expression construct was used to upregulate CISD2 and siRNA was used to knock down CISD2. Effects of manipulating CISD2 on autophagic markers LC3-II, beclin-1, Atg7 and p62 were determined using western blot analysis. Relative levels of (B) LC3-II, (C) beclin-1, (D) Atg7 and (E) p62 were measured using Image J software and normalized to GAPDH. Data are presented as the mean ± standard deviation from at least three independent experiments. *P<0.05 and **P<0.01, compared with the Ctl group. CISD2, CDGSH iron sulfur domain 2; si-CISD2, small interfering RNA targeting CISD2; Ctl, control; LC3-II, light chain 3 II; Atg7, autophagy related 7.
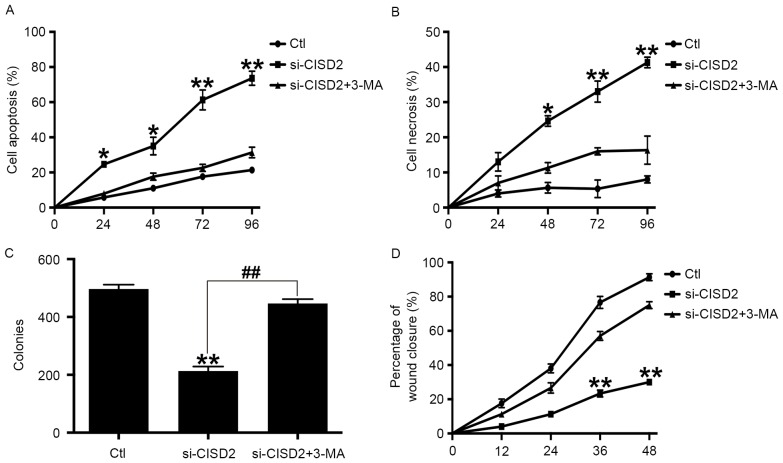
Inhibiting autophagy alleviates CISD2 silencing-induced glioma cell death
In order to confirm whether CISD2 silencing-induced glioma cell death was dependent on autophagy, the specific autophagy inhibitor, 3-MA, was used. Compared with the control group, si-CISD2 led to significant increases in cell apoptosis (Fig. 5A) and necrosis (Fig. 5B), however, these effects were eliminated by 3-MA. Consistently, the colony formation assay showed that si-CISD2 markedly reduced the mean number of colonies, compared with that in the control group, which was reversed by 3-MA (Fig. 5C). Finally, the wound-healing assay revealed that si-CISD significantly attenuated the invasive ability of U87 cells, which was also abrogated by 3-MA (Fig. 5D). Taken together, these data suggested that inhibiting autophagy significantly alleviated the CISD2 silencing-induced suppression of glioma cell proliferation and survival.
Figure 5.
Inhibiting autophagy alleviates CISD2 silencing-induced glioma cell death. 3-MA was used to inhibit autophagy. Flow cytometry revealed that 3-MA significantly eliminated the si-CISD2-induced increase in (A) cell apoptosis and (B) necrosis. (C) Colony formation assays showed that 3-MA reversed the si-CISD2-induced decrease in mean colony number. (D) Wound-healing assays revealed that 3-MA abrogated the si-CISD-induced compromise in the invasive ability of U87 cells. Data are presented as the mean ± standard deviation from at least three independent experiments. *P<0.05 and **P<0.01, compared with Ctl group and si-CISD2+3-MA group; ##P<0.01, compared with the indicated groups. CISD2, CDGSH iron sulfur domain 2; si-CISD2, small interfering RNA targeting CISD2; Ctl, control.
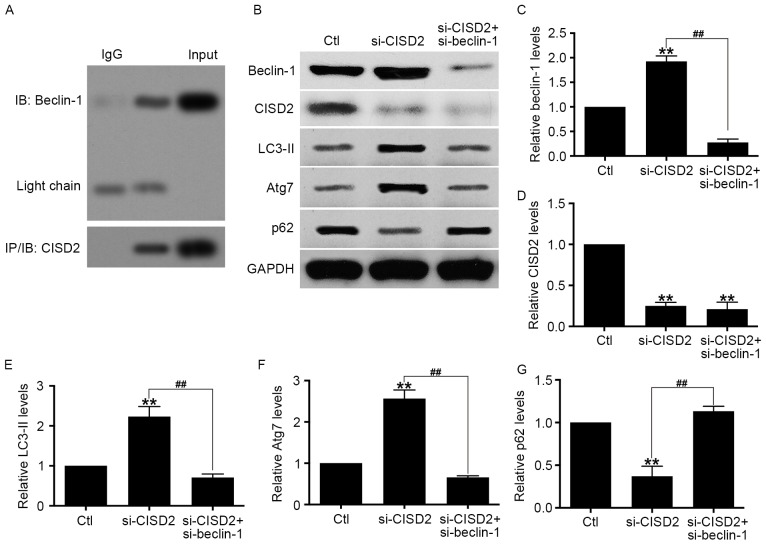
Activation of CISD2 silencing-induced autophagy is mediated by beclin-1
Previous studies have shown that beclin-1 is vital in the activation of autophagy (26). Therefore, the present study hypothesized that the enhanced autophagy by CISD2 silencing is dependent on beclin-1. The Co-IP assay showed that endogenous CISD2 was able to bind with beclin-1 (Fig. 6A). In addition, U87 cells with si-CISD2 and si-beclin-1 vectors were co-transfected, and the activity of autophagy activity was evaluated using western blot analysis. The results of the western blot analysis confirmed that CISD2 and beclin-1 were effectively knocked down (Fig. 6B-D). The data revealed that the downregulation of beclin-1 significantly eliminated the si-CISD2-induced increase in LC3-II and Atg7, and decrease in p62 in the U87 cells (Fig. 6E-G). Taken together, these data demonstrated that the CISD2 silencing-induced activation of autophagy was mediated by beclin-1.
Figure 6.
CISD2 silencing-induced activation of autophagy is mediated by beclin-1. (A) Co-immunoprecipitation assay showed the interaction between endogenous CISD2 and beclin-1. (B) U87 cells were co-transfected with si-CISD2 and si-beclin-1 plasmids, and the activity of autophagy was determine using western blot analysis. Relative levels of (C) beclin-1, (D) CISD2, (E) LC3-II, (F) Atg7 and (G) p62 were measured using Image J software and normalized to GAPDH. Data are presented as the mean ± standard deviation from at least three independent experiments. **P<0.01, compared with Ctl group; ##P<0.01, compared with the indicated groups. CISD2, CDGSH iron sulfur domain 2; si-CISD2, small interfering RNA targeting CISD2; LC3-II, light chain 3 II; Atg7, autophagy related 7; Ctl, control.
CISD2 is a direct target of miRNA-449a
Using TargetScan, the present study found that CISD2 was one of the potential targets of miR-449a. The predicted binding site of miR-449a with the CISD2 3′-UTR is shown in Fig. 7A. To confirm the interaction between miR-449a and CISD2, the CISD2 complementary sites, with or without mutations, were cloned into the 3′-UTR of the firefly luciferase gene and co-transfected with miR-449a mimics into U87 cells. The results revealed that the presence of miR-449a led to a significant reduction in the relative luciferase activity of the wild-type construct of the CISD2 3′-UTR in U87 cells. However, the mutant construct of the CISD2 3′-UTR eliminated the suppressive effect of miR-449a in the U87 cells (Fig. 7B). The above results demonstrated that CISD2 is a direct target of miR-449a.
Figure 7.
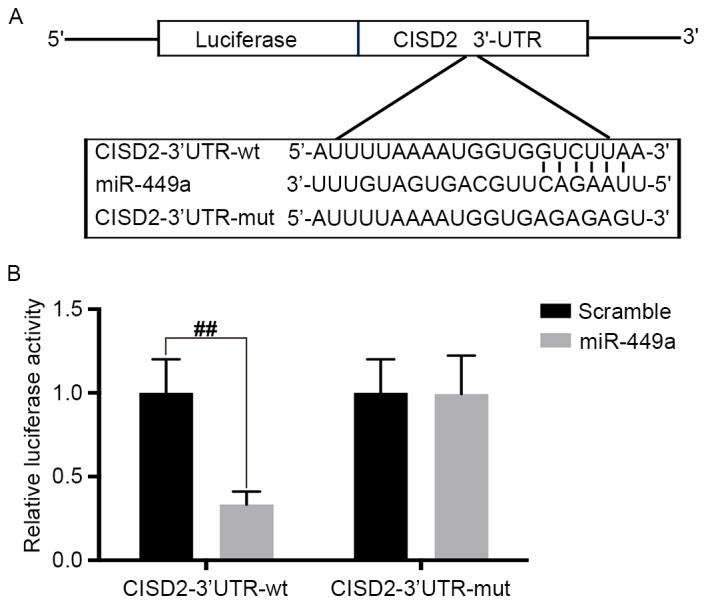
CISD2 is a direct target of miR-449a. (A) Predicted binding site of miR-449a with CISD2 3′-UTR. (B) pGL3 vector containing CISD2 wild-type 3′-UTR or mutated form was co-transfected with or without miR-449a mimic into U87 cells. At 48 h post-transfection, the luciferase activity was measured. Data are presented as the mean ± standard deviation from at least three independent experiments. ##P<0.01, compared with the indicated groups. CISD2, CDGSH iron sulfur domain 2; si-CISD2, small interfering RNA targeting CISD2; LC3-II, light chain 3 II; Atg7, autophagy related 7; 3′-UTR, 3′-untranslated region; miR, microRNA; wt, wild-type; mut, mutant.
Discussion
The present study provided the first evidence, to the best of our knowledge, that the mRNA and protein levels of CISD2 were upregulated in glioma tissues, compared with matched normal tissues. The inhibition of CISD2 via siRNA suppressed the proliferation and survival of glioma cells. Furthermore, the results demonstrated that the silencing of CISD2 inhibited the carcinogenesis of glioma cells in a xenograft model. Mechanistically, it was found that si-CISD2 significantly increased the activity of autophagy, whereas upregulated CISD2 markedly suppressed autophagy. Inhibiting autophagy with 3-MA significantly alleviated the CISD2 silencing-induced suppression of proliferation and survival of glioma cells. It was also found that endogenous CISD2 was able to bind with beclin-1. The downregulation of beclin-1 significantly eliminated the si-CISD2-induced increase of LC3-II and Atg7, and decrease of p62 in the U87 cells. This suggested that the activation of autophagy induced by silencing CISD2 was mediated by beclin-1. The results also revealed that CISD2 is a target of miR-449a.
Increasing evidence indicates that CISD2 is closely associated with multiple tumors. CISD2 is increased in human epithelial breast cancer cells, and suppressing CISD2 significantly inhibits tumor growth (11). CISD2 is upregulated and associated with poor prognosis in early stage cervical cancer (12), and CISD2 promotes the tumorigenesis and proliferation of gastric cancer cell through activating the AKT pathway (27). In hepatocellular carcinoma, the downregulation of CISD2 suppresses the proliferation of hepatoma cells (28). However, the role of CISD2 in glioma remains to be fully elucidated. To the best of our knowledge, the present study provided the first evidence that the mRNA and protein levels of CISD2 were upregulated in glioma tissues, compared with paired normal tissues. It was also demonstrated that si-CISD2 inhibited the proliferation, survival and invasion of glioma cells. These results suggested that CISD2 acts as an oncogene in glioma and that CISD2 may be a promising therapeutic target for glioma treatment.
The molecular mechanisms underlying the oncogenic role of CISD2 in tumors has been investigated previously. In breast cancer, CISD2 eliminates reactive oxygen and protects mitochondrial function (11). In gastric cancer, CISD2 activates the AKT signaling pathway to promote tumorigenesis (27). However, there remains no evidence demonstrating whether or not autophagy is involved in the tumorigenic role of CISD2. In the present study, it was found that si-CISD2 significantly increased the activity of autophagy, whereas upregulated levels of CISD2 markedly suppressed autophagy. Inhibiting autophagy with 3-MA significantly alleviated the CISD2 silencing-induced suppression of proliferation and survival of glioma cells. Therefore, the present study confirmed that CISD2 promoted tumorigenesis through inhibiting autophagic activity. However, whether or not the CISD2/autophagy pathway works in other tumors requires further investigation.
Studies have shown that beclin-1 is a mediator for the activation of autophagy in multiple conditions. In cerebral ischemic injury, beclin-1-mediated autophagy can alleviate neuronal damage (29). In addition, beclin-1-mediated autophagy contributes to the radiosensitivity of in pancreatic cancer cells (30). Beclin-1/autophagy is closely associated with prognosis in patients with non-metastatic renal cell carcinoma (31). Consistently, the present study showed that the downregulation of beclin-1 significantly eliminated the si-CISD2-induced increase of LC3-II and Atg7, and decrease of p62 in the U87 cells, suggesting that the activation of autophagy induced by CISD2 silencing was mediated by beclin-1. However, studies have shown that cyclin-dependent kinase 5 and heme oxygenase-1 are also mediators for the activation of autophagy (32–34). Consequently, it is necessary to investigate whether other mediators are involved in the si-CISD2-induced activation of autophagy.
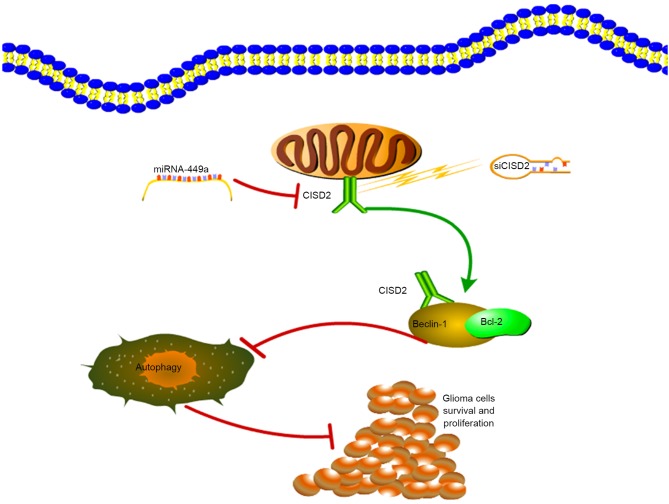
In conclusion, the present study demonstrated that CISD2 was increased in glioma samples, and was associated with poor prognosis and aggressive tumor behavior. The miR-449a/CISD2/beclin-1-mediated autophagy regulatory network contributed to the proliferation of glioma cells (Fig. 8). Therefore, targeting this pathway may be a promising strategy for glioma therapy.
Figure 8.
Schematic diagram of the contribution of the miR-449a/CISD2/beclin-1-mediated autophagy regulatory network to the proliferation of glioma cells. CISD2, CDGSH iron sulfur domain 2; si-CISD2, small interfering RNA targeting CISD2; miR, microRNA; Bcl-2, B-cell lymphoma 2.
Acknowledgements
The authors would like to thank Dr Haipeng Xu from Fudan University (Fudan, China) for proofreading of the manuscript.
References
- 1.Chen Y, Wu Y, Huang X, Qu P, Li G, Jin T, Xing J, He S. Leukocyte telomere length: A novel biomarker to predict the prognosis of glioma patients. J Cancer Res Clin Oncol. 2015;141:1739–1747. doi: 10.1007/s00432-015-1938-x. [DOI] [PubMed] [Google Scholar]
- 2.Wachsberger PR, Lawrence YR, Liu Y, Rice B, Feo N, Leiby B, Dicker AP. Hsp90 inhibition enhances PI-3 kinase inhibition and radiosensitivity in glioblastoma. J Cancer Res Clin Oncol. 2014;140:573–582. doi: 10.1007/s00432-014-1594-6. [DOI] [PubMed] [Google Scholar]
- 3.Gulati S, Jakola AS, Johannesen TB, Solheim O. Survival and treatment patterns of glioblastoma in the elderly: A population-based study. World Neurosurg. 2012;78:518–526. doi: 10.1016/j.wneu.2011.12.008. [DOI] [PubMed] [Google Scholar]
- 4.Fukushima T, Kawaguchi M, Yorita K, Tanaka H, Takeshima H, Umezawa K, Kataoka H. Antitumor effect of dehydroxymethylepoxyquinomicin, a small molecule inhibitor of nuclear factor-κB, on glioblastoma. Neuro Oncol. 2012;14:19–28. doi: 10.1093/neuonc/nor168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lin J, Zhang L, Lai S, Ye K. Structure and molecular evolution of CDGSH iron-sulfur domains. PLoS One. 2011;6:e24790. doi: 10.1371/journal.pone.0024790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wu CY, Chen YF, Wang CH, Kao CH, Zhuang HW, Chen CC, Chen LK, Kirby R, Wei YH, Tsai SF, Tsai TF. A persistent level of Cisd2 extends healthy lifespan and delays aging in mice. Hum Mol Genet. 2012;21:3956–3968. doi: 10.1093/hmg/dds210. [DOI] [PubMed] [Google Scholar]
- 7.Wang CH, Kao CH, Chen YF, Wei YH, Tsai TF. Cisd2 mediates lifespan: Is there an interconnection among Ca2+ homeostasis, autophagy, and lifespan? Free Radic Res. 2014;48:1–1114. doi: 10.3109/10715762.2014.936431. [DOI] [PubMed] [Google Scholar]
- 8.Tsai PH, Chien Y, Chuang JH, Chou SJ, Chien CH, Lai YH, Li HY, Ko YL, Chang YL, Wang CY, et al. Dysregulation of mitochondrial functions and osteogenic differentiation in cisd2-deficient murine induced pluripotent stem cells. Stem Cells Dev. 2015;24:2561–2576. doi: 10.1089/scd.2015.0066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chang NC, Nguyen M, Bourdon J, Risse PA, Martin J, Danialou G, Rizzuto R, Petrof BJ, Shore GC. Bcl-2-associated autophagy regulator Naf-1 required for maintenance of skeletal muscle. Hum Mol Genet. 2012;21:2277–2287. doi: 10.1093/hmg/dds048. [DOI] [PubMed] [Google Scholar]
- 10.Chen YF, Kao CH, Chen YT, Wang CH, Wu CY, Tsai CY, Liu FC, Yang CW, Wei YH, Hsu MT, et al. Cisd2 deficiency drives premature aging and causes mitochondria-mediated defects in mice. Genes Dev. 2009;23:1183–1194. doi: 10.1101/gad.1779509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sohn YS, Tamir S, Song L, Michaeli D, Matouk I, Conlan AR, Harir Y, Holt SH, Shulaev V, Paddock ML, et al. NAF-1 and mitoNEET are central to human breast cancer proliferation by maintaining mitochondrial homeostasis and promoting tumor growth; Proc Natl Acad Sci USA; 2013; pp. 14676–14681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu L, Xia M, Wang J, Zhang W, Zhang Y, He M. CISD2 expression is a novel marker correlating with pelvic lymph node metastasis and prognosis in patients with early-stage cervical cancer. Med Oncol. 2014;31:183. doi: 10.1007/s12032-014-0183-5. [DOI] [PubMed] [Google Scholar]
- 13.Wang B, Cai Z, Tao K, Zeng W, Lu F, Yang R, Feng D, Gao G, Yang Q. Essential control of mitochondrial morphology and function by chaperone-mediated autophagy through degradation of PARK7. Autophagy. 2016;12:1215–1228. doi: 10.1080/15548627.2016.1179401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wei Y, Pattingre S, Sinha S, Bassik M, Levine B. JNK1-mediated phosphorylation of Bcl-2 regulates starvation-induced autophagy. Mol Cell. 2008;30:678–688. doi: 10.1016/j.molcel.2008.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang JD, Cao YL, Li Q, Yang YP, Jin M, Chen D, Wang F, Wang GH, Qin ZH, Hu LF, Liu CF. A pivotal role of FOS-mediated BECN1/Beclin 1 upregulation in dopamine D2 and D3 receptor agonist-induced autophagy activation. Autophagy. 2015;11:2057–2073. doi: 10.1080/15548627.2015.1100930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Choi J, Jung W, Koo JS. Expression of autophagy-related markers beclin-1, light chain 3A, light chain 3B and p62 according to the molecular subtype of breast cancer. Histopathology. 2013;62:275–286. doi: 10.1111/his.12002. [DOI] [PubMed] [Google Scholar]
- 17.Komatsu M, Ichimura Y. Physiological significance of selective degradation of p62 by autophagy. FEBS Lett. 2010;584:1374–1378. doi: 10.1016/j.febslet.2010.02.017. [DOI] [PubMed] [Google Scholar]
- 18.White E. Deconvoluting the context-dependent role for autophagy in cancer. Nat Rev Cancer. 2012;12:401–410. doi: 10.1038/nrc3262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.He WS, Dai XF, Jin M, Liu CW, Rent JH. Hypoxia-induced autophagy confers resistance of breast cancer cells to ionizing radiation. Oncol Res. 2012;20:251–258. doi: 10.3727/096504013X13589503483012. [DOI] [PubMed] [Google Scholar]
- 20.Mohapatra P, Preet R, Das D, Satapathy SR, Choudhuri T, Wyatt MD, Kundu CN. Quinacrine-mediated autophagy and apoptosis in colon cancer cells is through a p53- and p21-dependent mechanism. Oncol Res. 2012;20:81–91. doi: 10.3727/096504012X13473664562628. [DOI] [PubMed] [Google Scholar]
- 21.Sasazawa Y, Sato N, Umezawa K, Simizu S. Conophylline protects cells in cellular models of neurodegenerative diseases by inducing mammalian target of rapamycin (mTOR)-independent autophagy. J Biol Chem. 2015;290:6168–6178. doi: 10.1074/jbc.M114.606293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brassesco MS, Pezuk JA, Morales AG, de Oliveira JC, Valera ET, da Silva GN, de Oliveira HF, Scrideli CA, Umezawa K, Tone LG. Cytostatic in vitro effects of DTCM-glutarimide on bladder carcinoma cells. Asian Pac J Cancer Prev. 2012;13:1957–1962. doi: 10.7314/APJCP.2012.13.5.1957. [DOI] [PubMed] [Google Scholar]
- 23.Allen M, Bjerke M, Edlund H, Nelander S, Westermark B. Origin of the U87MG glioma cell line: Good news and bad news. Sci Transl Med. 2016;8:354re3. doi: 10.1126/scitranslmed.aaf6853. [DOI] [PubMed] [Google Scholar]
- 24.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 25.Yuan J, Zhang Y, Sheng Y, Fu X, Cheng H, Zhou R. MYBL2 guides autophagy suppressor VDAC2 in the developing ovary to inhibit autophagy through a complex of VDAC2-BECN1-BCL2L1 in mammals. Autophagy. 2015;11:1081–1098. doi: 10.1080/15548627.2015.1040970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Carchman EH, Rao J, Loughran PA, Rosengart MR, Zuckerbraun BS. Heme oxygenase-1-mediated autophagy protects against hepatocyte cell death and hepatic injury from infection/sepsis in mice. Hepatology. 2011;53:2053–2062. doi: 10.1002/hep.24324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang L, Ouyang F, Liu X, Wu S, Wu HM, Xu Y, Wang B, Zhu J, Xu X, Zhang L. Overexpressed CISD2 has prognostic value in human gastric cancer and promotes gastric cancer cell proliferation and tumorigenesis via AKT signaling pathway. Oncotarget. 2016;7:3791–3805. doi: 10.18632/oncotarget.6302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen B, Shen S, Wu J, Hua Y, Kuang M, Li S, Peng B. CISD2 associated with proliferation indicates negative prognosis in patients with hepatocellular carcinoma. Int J Clin Exp Pathol. 2015;8:13725–13738. [PMC free article] [PubMed] [Google Scholar]
- 29.Wang P, Liang J, Li Y, Li J, Yang X, Zhang X, Han S, Li S, Li J. Down-regulation of miRNA-30a alleviates cerebral ischemic injury through enhancing beclin 1-mediated autophagy. Neurochem Res. 2014;39:1279–1291. doi: 10.1007/s11064-014-1310-6. [DOI] [PubMed] [Google Scholar]
- 30.Zhang X, Shi H, Lin S, Ba M, Cui S. MicroRNA-216a enhances the radiosensitivity of pancreatic cancer cells by inhibiting beclin-1-mediated autophagy. Oncol Rep. 2015;34:1557–1564. doi: 10.3892/or.2015.4078. [DOI] [PubMed] [Google Scholar]
- 31.Nishikawa M, Miyake H, Liu B, Fujisawa M. Expression pattern of autophagy-related markers in non-metastatic clear cell renal cell carcinoma: Association with disease recurrence following radical nephrectomy. J Cancer Res Clin Oncol. 2015;141:1585–1591. doi: 10.1007/s00432-015-1923-4. [DOI] [PubMed] [Google Scholar]
- 32.Wong AS, Lee RH, Cheung AY, Yeung PK, Chung SK, Cheung ZH, Ip NY. Cdk5-mediated phosphorylation of endophilin B1 is required for induced autophagy in models of Parkinson's disease. Nat Cell Biol. 2011;13:568–579. doi: 10.1038/ncb0611-734d. [DOI] [PubMed] [Google Scholar]
- 33.Wen Z, Shu Y, Gao C, Wang X, Qi G, Zhang P, Li M, Shi J, Tian B. CDK5-mediated phosphorylation and autophagy of RKIP regulate neuronal death in Parkinson's disease. Neurobiol Aging. 2014;35:2870–2880. doi: 10.1016/j.neurobiolaging.2014.05.034. [DOI] [PubMed] [Google Scholar]
- 34.Wang Y, Shen J, Xiong X, Xu Y, Zhang H, Huang C, Tian Y, Jiao C, Wang X, Li X. Remote ischemic preconditioning protects against liver ischemia-reperfusion injury via heme oxygenase-1-induced autophagy. PLoS One. 2014;9:e98834. doi: 10.1371/journal.pone.0098834. [DOI] [PMC free article] [PubMed] [Google Scholar]