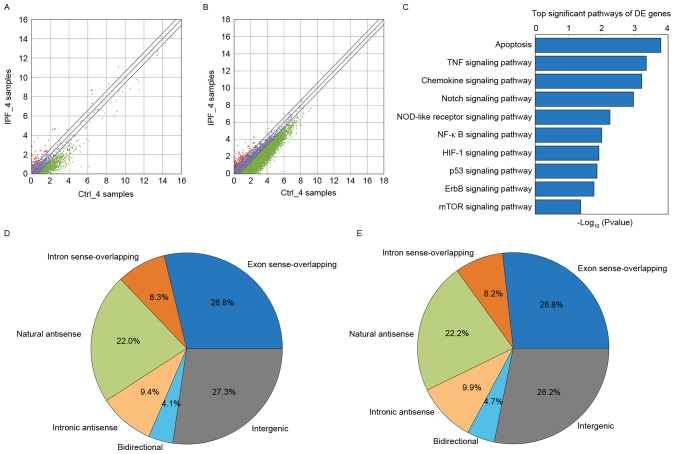
Figure 1.
Bioinformatics analysis. (A) Differentially-expressed lncRNAs. A total of 440 lncRNAs that were upregulated and 1,376 lncRNAs that were downregulated in the IPF group. Red indicates high relative expression and green indicates low relative expression. Purple indicates no significant change. (B) Differentially-expressed mRNAs. There were 361 mRNAs that were upregulated and 1,124 mRNAs that were downregulated in the IPF group. Red indicates high relative expression and green indicates low relative expression. (C) Target enriched pathways. The abscissa represents the enrichment score. The mTOR signaling pathway was one of the significant pathways, according to the pathway analysis. (D) According to their position in the genome, which is associated with protein-coding genes, lncRNAs may be divided into six types: Exon sense-overlapping; intronic antisense; natural antisense; intergenic; bidirectional; and intron sense-overlapping. Natural antisense represented 22% of the total in the IPF group. (E) Natural antisense accounted for 22.2% in the control group. lncRNA, long noncoding RNA; IPF, idiopathic pulmonary fibrosis; mTOR, protein kinase mTOR; Ctrl, control.