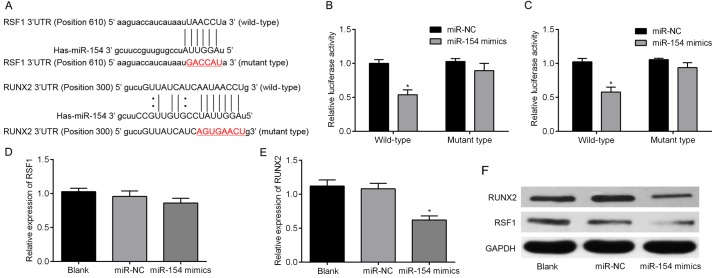
Figure 2.
miR-154 directly targets RSF1 and RUNX2 in BC cells. (A) Several bioinformatics tools were used to predict miR-154 binding sites. This identified putative sites in the 3′-UTRs of the RSF1 and RUNX2 genes. (B) The relative luciferase activities of cells co-transfected with either wild WT or MUT 3′-UTR constructs containing RSF1 and either miR-154 mimics or miR-NC. (C) The relative luciferase activity of cells transfected with WT/MUT 3′-UTR constructs containing RUNX2 and either miR-154 mimics or miR-NC. (D) The expression levels of RSF1 mRNA in T24 cells transfected with miR-154 or miR-NC. (E) RUNX2 was significantly lower in cells transfected miR-154 mimics compared to the miR-NC group, assessed by RT-qPCR. (F) miR-154 expression affected both RSF1 and RUNX2 protein expression in T24 cells; *P<0.05 vs. the miR-NC group. RSF1, remodeling and spacing factor 1; RUNX2, runt-related transcription factor 2; BC, bladder cancer; Wt, wild-type; MUT, mutant-type.