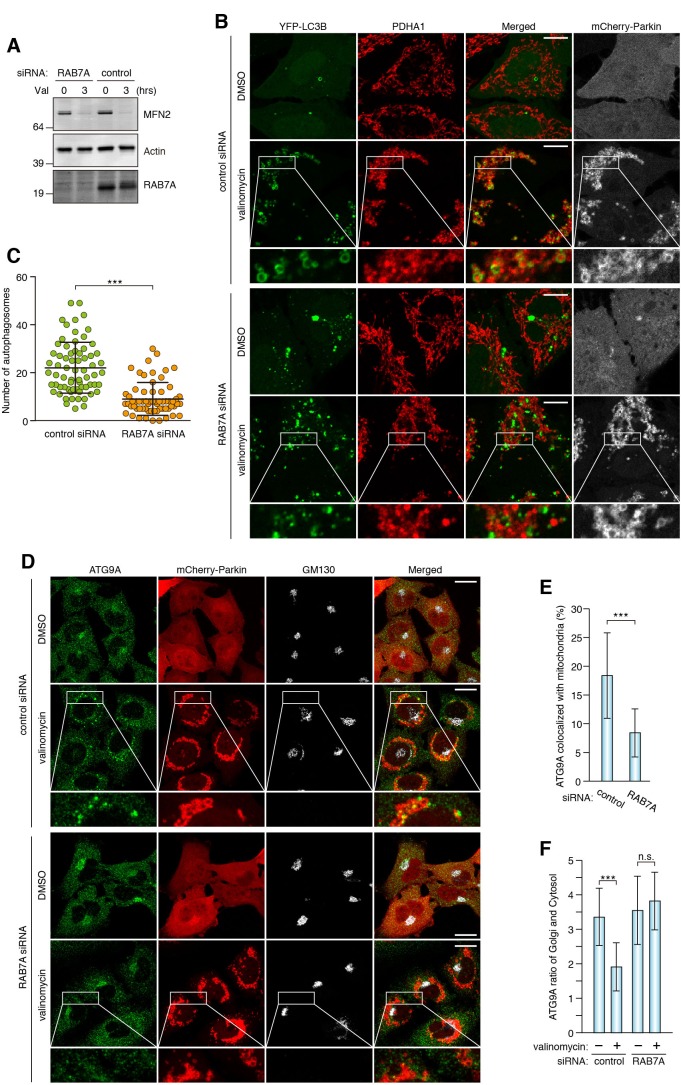
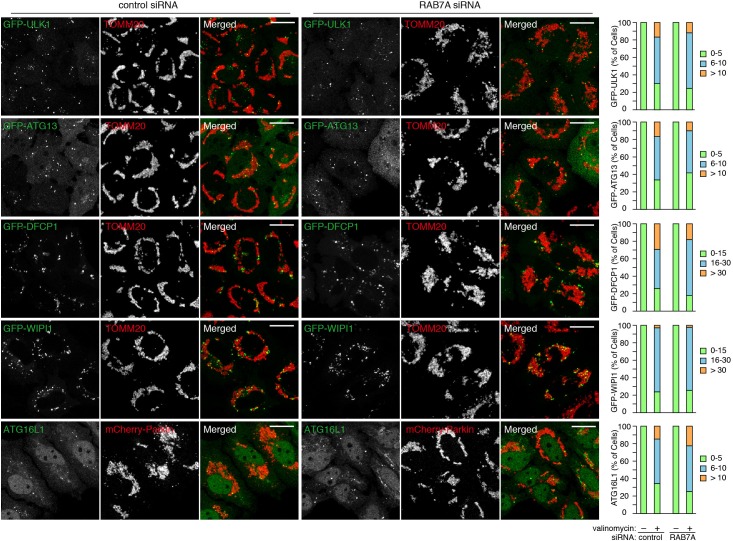
Figure 3. RAB7A is required for ATG9A recruitment to damaged mitochondria and encapsulation by autophagic membranes.
(A) siRNA-treated HeLa cells stably expressing mCherry-Parkin were treated with DMSO or valinomycin (Val) for 3 hr. Total cell lysates were analyzed by immunoblotting. (B) siRNA-treated HeLa cells stably expressing mCherry-Parkin and YFP-LC3B were treated with DMSO or valinomycin for 3 hr. The fixed cells were subjected to immunostaining. Images are displayed as z-stacks of five confocal slices. The magnified pictures of the cells treated with valinomycin were shown. Bars, 10 μm. (C) The number of autophagosomes containing PDHA1 inside in each cell was counted. Error bars represent mean ±SE of at least two independent experiments. Statistical differences were determined by student’s t-test. ***p<0.001. (D) The fixed cells as in (A) were subjected to immunostaining. Images are displayed as z-stacks of five confocal slices. Magnified images are shown for cells treated with valinomycin. Bars, 20 μm. (E) Quantification of ATG9A recruitment to damaged mitochondria in (D). Overlapped ATG9A signals with mitochondria-localized mCherry-Parkin per total ATG9A signals were measured. Total ATG9A signal in each cell set to 100%. Error bars represent mean ±SE. Cells from at least two independent experiments were quantified. Statistical difference was determined by student’s t-test. ***p<0.001. (F) Quantification of ATG9A localization on Golgi apparatus (see the Materials and methods for the detail). Error bars represent mean ±SE. Cells from at least two independent experiments were quantified. Statistical difference was determined by student’s t-test ***p<0.001; n.s., not significant.