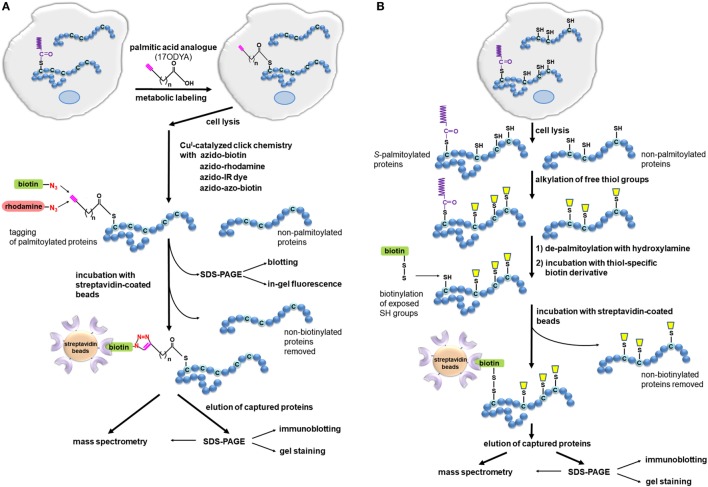
Figure 2.
Detection of S-palmitoylated proteins using click chemistry and acyl-biotin exchange (ABE). (A) Click chemistry-based method. Cells are metabolically labeled with an alkyne-functionalized palmitic acid analog, such as 17-octadecynoic acid (17ODYA), and after cell lysis, the click reaction is conducted with azido-tagged biotin or fluorescent probes allowing enrichment and detection of labeled proteins in various ways. Biotinylated proteins can be bound on a streptavidin resin and then released using, e.g., high concentrations of urea and SDS (108). When a cleavable derivative of biotin, azido-azo-biotin, is used the labeled proteins are eluted from streptavidin beads with sodium dithionite, which cleaves the diazobenzene moiety in the linker arm of azido-azo-biotin, and analyzed by mass spectrometry or immunoblotting (109). (B) ABE method. Cells or tissues are lysed, free thiol groups of proteins are blocked by alkylation, and palmitoyl moieties are released with hydroxylamine. The newly exposed protein thiol groups are subjected to labeling with biotin-HPDP allowing selective binding, elution, and analysis of the originally S-palmitoylated proteins. The proteins can also be captured without biotinylation through a direct interaction of their thiol residues with a thiol-reactive resin (acyl-RAC technique).