Abstract
Please cite this paper as: Bodle et al. (2013) Development of an enzyme‐linked immunoassay for the quantitation of influenza haemagglutinin: an alternative method to single radial immunodiffusion. Influenza and Other Respiratory Viruses 7(2) 191–200.
Background The current method used to measure haemagglutinin (HA) content for influenza vaccine formulation, single radial immunodiffusion (SRID), is lengthy and relies on the availability of matched standardised homologous reagents. The 2009 influenza pandemic highlighted the need to develop alternate assays that are able to rapidly quantitate HA antigen for vaccine formulation.
Objectives The aim of this work was to develop an enzyme‐linked immunoassay (EIA) for the rapid quantitation of H1, H3, H5 and B influenza HA antigens.
Methods Monoclonal antibodies (mAbs) selected for haemagglutination inhibition (HAI) activity were conjugated with horseradish peroxidase and used to establish a capture–detection EIA for the quantitation of HA antigen. Results were compared with the appropriate reference SRID assays to investigate assay performance and utility.
Results Quantitation of HA antigen by EIA correlated well with current reference SRID assays. EIA results showed equivalent precision and exhibited a similar capacity to detect HA antigen in virus samples that had been used in either stability or splitting studies, or subjected to physical or chemical stresses. EIA exhibited greater sensitivity than SRID and has the potential to be used in high‐throughput applications.
Conclusions We demonstrated the utility of EIA as a suitable alternative to SRID for HA antigen quantitation and stability assessment. This approach would lead to earlier availability of both seasonal and pandemic vaccines, because of the extended cross‐reactivity of reagents.
Keywords: Enzyme‐linked immunoassay, haemagglutinin, influenza vaccine, single radial immunodiffusion, vaccine stability
Introduction
Vaccines are used to minimise the worldwide burden of influenza, a major disease that kills between 250 000–500 000 people annually. 1 Haemagglutinin (HA) presents the major target for virus‐neutralising antibodies against influenza and is considered the most important component for protection. 2 , 3 To elicit an adequate neutralising antibody response in recipients, influenza vaccines are formulated to a specific HA antigen content.
Regulatory guidelines specify the use of an immunological assay for vaccine HA antigen quantitation, and to date, the single radial immunodiffusion (SRID) assay has been widely employed for this purpose. 4 , 5 SRID utilises strain‐specific sheep polyclonal antiserum raised against purified preparations of HA antigen, and quantitation of HA antigen is achieved using whole‐virus standards with known HA antigen content. Whilst SRID has been used during influenza vaccine manufacture for many decades, it has several limitations. The SRID assay is time consuming, the process is unable to be automated and the assay has relatively poor accuracy and sensitivity. 6 , 7 , 8 , 9 , 10 Furthermore, SRID generally requires the use of strain‐specific reagents, a reliance that can delay the development and release of vaccine. An alternative assay that detects immunologically relevant antigens, yet reduces the reliance on homologous reagents and has the capacity for high‐throughput automation is highly desirable.
High‐performance liquid chromatography (HPLC) and mass spectrometry have been proposed as alternate assays for HA quantitation; however, these approaches require denaturation of the HA antigen. 7 , 10 They are unable to assess the immune relevance of the antigen or vaccine stability and thus are not suitable for influenza vaccine manufacture.
Several groups have examined enzyme‐linked immunoassay (EIA) as an alternative for HA antigen quantitation; 6 , 9 however, these studies did not examine EIA quantitation of immunologically relevant HA antigen or quantitation in comparison to SRID.
Here, we report the development of multiple subtype‐specific EIAs for the quantitation of HA antigen. We assessed the correlation of these assays with corresponding SRID assays and demonstrated that these assays can detect relevant influenza HA antigens across H1N1, pandemic H1N1 (2009), H3N2, H5N1 and B subtypes. Significantly, many of our assays could accurately quantitate HA antigen across multiple drifted strains of influenza. We also demonstrated increased sensitivity for this approach and comparable precision and stability analysis in response to physical and chemical stresses.
Materials and methods
Antibodies
Monoclonal antibodies were produced as previously described 11 using density gradient–purified whole influenza virus propagated in embryonated hens’ eggs as immunogens. Antibodies were raised against the influenza strains A/Hiroshima/52/05 (H3N2; HIR3.8G11.1B5), A/Singapore/37/09 (H3N2; SIN3.9G11.2B6), A/Brisbane/59/07 (H1N1; ABR4.1D5.1B11), A/California/07/09 (H1N1; CAL2.5C6.1E3), A/Indonesia/05/05 (H5N1; ML3.4C9.1B2), A/Vietnam/1194/04 (H5N1; VI8.5C2.1B10), B/Panama/45/90 (PM10.2D7.2F11.1G6), B/Malaysia/2506/04 (MIA1.8E3.1E9), B/Brisbane/60/08 (BBR3.10E10.1C3) and B/Wisconsin/1/10 (BWIS141.9B3.2D8). The resulting hybridomas were screened and selected on the basis of a positive haemagglutination inhibition (HAI) test. 12
For EIA, mAbs were concentrated using a Mini‐Perm system (Greiner Bio‐One, Kremsmuenster, Austria) and, where required, purified by buffer exchange using 0·1 m carbonate buffer (pH 9·5). For the sandwich EIA, mAbs were conjugated with horseradish peroxidase (HRP) according to manufacturers’ specifications (Lightning Link; Innova Biosciences, Cambridge, UK).
Sheep polyclonal anti‐HA antibodies and inactivated sucrose gradient‐purified influenza, standardised for HA potency, were obtained from either the Therapeutic Goods Administration (TGA; Woden, ACT, Australia), the National Institute for Biological Standards and Control (NIBSC; Potters Bar, Hertfordshire, UK), or the Centre for Biological Evaluation and Research (CBER; Rockville, MD, USA). These antisera were raised against A/Indonesia/05/05 (CBER), A/Vietnam/1194/04 (NIBSC), A/Wisconsin/67/05 (H3N2, NIBSC), A/Brisbane/10/07 (H3N2, TGA), A/Uruguay/716/07 (H3N2, TGA), A/Brisbane/59/07 (NIBSC), B/Brisbane/3/07 (TGA), B/Florida/4/06 (NIBSC) and B/Malaysia/2506/04 (NIBSC), B/Brisbane/60/08 (TGA), A/Victoria/210/09 (H3N2, TGA), A/New York/55/04 (H3N2, NIBSC) and A/New Caledonia/20/99 (H1N1, NIBSC).
Influenza viruses
To confirm reactivity, mAbs were screened against viruses selected from the following panels: H1N1: A/Puerto Rico/8/34, A/New Caledonia/20/99, A/Solomon Islands/3/06, A/Brisbane/59/07, A/Guam/1/07, A/Pennsylvania/8/08, A/Perth/21/09. H5N1: A/Vietnam/1194/04, A/Indonesia/05/05. H3N2: A/Northern Territory/60/68, A/Port Chalmers/1/73, A/England/864/75, A/Bangkok/1/79, A/Shanghai/31/80, A/New York/55/04, A/Philippines/2/82, A/Victoria/5/84, A/Mississippi/1/85, A/Shanghai/11/87, A/Beijing/353/81, A/Guangdong/25/93, A/Sydney/5/97, A/Moscow/10/99, A/Panama/2007/99, A/Wyoming/3/03, A/Wellington/1/04, A/Nepal/921/06, A/New York/55/04, A/Wisconsin/67/05, A/Hiroshima/52/05, A/Brisbane/10/07, A/Uruguay/716/07, A/Victoria/210/09, A/Victoria/208/09, A/Alaska/05/10 and A/Victoria/563/10. B strain: B/Great Lakes/1739/54, B/Hong Kong/5/72, B/Harbin/07/94, B/Yamanashi/116/98, B/Victoria/504/00, B/Jiangsu/10/03 and B/Malaysia/2506/04, B/Yamagata/16/88, B/Panama/45/90, B/Florida/4/06, B/Brisbane/3/07, B/Bangladesh/3333/07, B/England/145/08, B/Brisbane/60/08, B/Brisbane/46/08, B/Hubei‐Wujiagang/158/09 and B/Wisconsin/1/10. Viruses for testing were propagated in embryonated hens’ eggs.
Trivalent influenza vaccine (TIV)
CSL TIV 2007RF (CSL Ltd, Parkville, VIC, Australia) contained strains A/New Caledonia/20/99 (H1N1), A/Wisconsin/67/05 (H3N2) and B/Malaysia/2506/04. CSL TIV 2007 (CSL Ltd) contained the strains A/Solomon Island/3/06 (H1N1), A/Wisconsin/67/05 (H3N2) and B/Malaysia/2506/04. CSL TIV 2007/2008 (CSL Ltd) contained the strains A/Solomon Island/3/06 (H1N1), A/Brisbane/10/07 (H3N2) and B/Brisbane/3/07.
Monovalent pooled harvest (MPH)
Samples were collected for EIA and SRID testing during the MPH stage of vaccine manufacturing process. To produce MPH, purified and concentrated virus preparations, which have been inactivated using β‐propiolactone, are split using sodium taurodeoxycholate and undergo filtration to remove detergent using proprietary methods (CSL Ltd). Monovalent pooled harvest preparations of the following viruses were used: A/Brisbane/59/07 (H1N1), A/Brisbane/10/07 (H3N2), A/Uruguay/716/07 (H3N2), A/Victoria/210/09 (H3N2), B/Brisbane/3/07, B/Florida/4/06 and B/Brisbane/60/08. MPH preparations of known HA antigen concentration, measured using SRID, were used as standards.
Inactivated sucrose gradient–purified influenza standardised for HA potency, used as a standard for SRID, was obtained from the TGA, NIBSC or CBER as described for sheep polyclonal antisera.
Enzyme‐linked immunoassay
Antigen capture and detection using native and conjugated mAbs was used to detect influenza HA antigen as previously described. 9 Briefly, microtitre plates (Nunc, Rochester, NY, USA) were coated with unconjugated mAb diluted in 0·05 m carbonate buffer and incubated at room temperature (RT) overnight. Plates were blocked for non‐specific binding using 1% (w/v) sodium casein solution in calcium‐ and magnesium‐free PBS (PBS−). Serial twofold dilutions of HA samples were added to the plate, in duplicate, and incubated for 1 hour at RT. Plates were washed with PBS− containing 0·05% (v/v) Tween‐20 (Sigma‐Aldrich, St. Louis, MO, USA) and incubated with HRP‐conjugated mAb for 1 hour at RT, washed and then incubated with peroxidase substrate (KPL, Gaithersburg, MD, USA) as recommended. Substrate development was ceased by the addition of 0·55 m H2SO4. The optical density (OD) was measured using a plate reader (Tecan, Mannedorf, Switzerland) with filters suitable to detect 450 nm. The concentration of HA was calculated by comparison with standards by 4‐parameter statistics using Magellan version 4 software (Tecan). Optimisation of coating and detection for each EIA was performed by titration of unconjugated and conjugated mAbs, respectively, with fixed concentrations of purified inactivated virus. 13 The optimum mAb concentration was calculated to be the lowest level of antibody that gave maximum OD at 450 nm. Haemagglutinin samples were considered to be positive for the presence of HA if the OD at 450 nm was above a threshold of 4 standard deviations above the mean background reading.
Single radial immunodiffusion assay
SRID assays were performed as previously described. 14 Briefly, reference and test antigen materials were treated with PBS− containing 1% (w/v) Zwittergent solution (Calbiochem, Darmstadt, Germany) and added to duplicate wells of agarose gels containing polyclonal antiserum diluted as recommended by the manufacturer. Gels were incubated for 72 hours in humidified chambers, dried onto glass plates and stained with Coomassie Brilliant Blue R‐250 (Sigma). Circular zones of antigen–antibody precipitation were measured, and HA concentration was calculated by the parallel line bioassay method in comparison with IZP standards, 15 and test validity was confirmed using a ‘g’ test (g ≤ 0·061). 16
Stability‐indicating properties of EIA and SRID
Trivalent vaccine material was subjected to the following stress tests: heat (60°C for 10, 30 or 120 minute); oxidation [0·001, 0·01, 1 or 5% (v/v) hydrogen peroxide at RT for 1 hour]; and pH (adjusted to pH 2, 3, 5 or 6 with HCl, or pH 8, 9 or 10 with NaOH, for 1 hour at RT and subsequently neutralised to pH 7·1). The HA concentration of the stressed material was then measured using the relevant EIA or SRID assay.
Statistical analysis
Differences in HA concentrations determined by the EIA and SRID assays were compared between groups in two‐sample Student’s t‐tests. P < 0·05 was taken as the level of statistical significance.
Results
EIA development
Monoclonal antibodies were raised against six influenza A strains [A/Hiroshima/52/05 (H3N2), A/Singapore/37/09 (H3N2), A/Brisbane/59/07 (H1N1), A/California/07/09 (H1N1), A/Indonesia/5/05 (H5N1) and A/Vietnam/1194/04 (H5N1)] and four influenza B strains [B/Panama/45/90, B/Malaysia/2506/04, B/Brisbane/60/08 and B/Wisconsin/1/10] and were confirmed to be specific for HA by HAI testing (data not shown). To determine subtype specificity, each mAb was screened against a panel of viruses by EIA. All mAbs were found to be highly subtype specific (Table 1).
Table 1.
EIA reactivity for influenza subtypes H1N1, H1N1 2009 Pandemic (H1N1 Pan), H3N2, H5N1 and influenza B strains (B)
| A/Bri | A/Cal | A/Hir | A/Sin | A/Vie | A/Ind | B/Mal | B/Bri | B/Pa | B/Wis | |
|---|---|---|---|---|---|---|---|---|---|---|
| H1N1 | ||||||||||
| A/Puerto Rico/8/34 | − | − | − | − | − | − | − | − | − | − |
| A/New Caledonia/20/99 | − | − | − | − | − | − | − | − | − | − |
| A/Solomon Island/3/06 | + | − | − | − | − | − | − | − | − | − |
| A/Fukushima/141/06 | + | − | − | − | − | − | − | − | − | − |
| A/Brisbane/59/07 | + | − | − | − | − | − | − | − | − | − |
| A/Guam/1/07 | − | − | − | − | − | − | − | − | − | − |
| A/Pennsylvania/8/08 | − | − | − | − | − | − | − | − | − | − |
| A/Perth/21/09 | − | − | − | − | − | − | − | − | − | − |
| H1N1 Pan | ||||||||||
| A/California/07/09 | − | + | − | − | − | − | − | − | − | − |
| A/Brisbane/10/10 | − | + | − | − | − | − | − | − | − | − |
| A/Brisbane/70/11 | − | + | − | − | − | − | − | − | − | − |
| A/Victoria/502/10 | − | + | − | − | − | − | − | − | − | − |
| H3N2 | ||||||||||
| A/Northern Territory/60/68 | NT | NT | − | NT | NT | NT | NT | NT | NT | NT |
| A/Port Chalmers/1/73 | NT | NT | − | NT | NT | NT | NT | NT | NT | NT |
| A/England/864/75 | NT | NT | − | NT | NT | NT | NT | NT | NT | NT |
| A/Texas/1/77 | − | − | − | − | − | − | − | − | − | − |
| A/Bangkok/1/79 | NT | NT | − | NT | NT | NT | NT | NT | NT | NT |
| A/Shanghai/31/80 | NT | NT | − | NT | NT | NT | NT | NT | NT | NT |
| A/Philippines/2/82 | NT | NT | − | NT | NT | NT | NT | NT | NT | NT |
| A/Victoria/5/84 | NT | NT | − | NT | NT | NT | NT | NT | NT | NT |
| A/Mississippi/1/85 | NT | NT | − | NT | NT | NT | NT | NT | NT | NT |
| A/Shanghai/11/87 | NT | NT | − | NT | NT | NT | NT | NT | NT | NT |
| A/Beijing/353/81 | NT | NT | − | NT | NT | NT | NT | NT | NT | NT |
| A/Guangdong/25/93 | NT | NT | − | NT | NT | NT | NT | NT | NT | NT |
| A/Sydney/5/97 | NT | NT | − | NT | NT | NT | NT | NT | NT | NT |
| A/Moscow/10/99 | NT | NT | − | NT | NT | NT | NT | NT | NT | NT |
| A/Panama/2007/99 | − | − | − | − | − | − | − | − | − | − |
| A/Wyoming/3/03 | − | − | + | − | − | − | − | − | − | − |
| A/Wellington/1/04 | − | − | + | − | − | − | − | − | − | − |
| A/Hiroshima/52/05 | − | − | + | − | − | − | − | − | − | − |
| A/Nepal/921/06 | − | − | + | + | − | − | − | − | − | − |
| A/Brisbane/10/07 | − | − | + | − | − | − | − | − | − | − |
| A/Uruguay/716/07 | − | − | + | − | − | − | − | − | − | − |
| A/Brisbane/24/08 | − | − | + | − | − | − | − | − | − | − |
| A/Wisconsin/15/09 | − | − | − | + | − | − | − | − | − | − |
| A/Victoria/210/09 | − | − | − | + | − | − | − | − | − | − |
| A/Victoria/208/09 | − | − | − | + | − | − | − | − | − | − |
| A/Alaska/05/10 | − | − | − | + | − | − | − | − | − | − |
| A/Victoria/563/10 | − | − | − | + | − | − | − | − | − | − |
| H5N1 | ||||||||||
| A/Vietnam/1194/04 | − | − | − | − | + | − | − | − | − | − |
| A/Indonesia/05/05 | − | − | − | − | + | + | − | − | − | − |
| B | ||||||||||
| B/Great Lakes/1739/54 | − | − | − | − | − | − | − | − | − | − |
| B/Hong Kong/5/72 | − | − | − | − | − | − | − | − | − | − |
| B/Yamagata/16/88 | − | − | − | − | − | − | − | − | + | − |
| B/Panama/45/90 | − | − | − | − | − | − | − | − | + | − |
| B/Harbin/07/94 | − | − | − | − | − | − | − | − | − | + |
| B/Yamanashi/166/98 | − | − | − | − | − | − | − | − | − | + |
| B/Victoria/504/00 | − | − | − | − | − | − | − | − | − | + |
| B/Jiangsu/10/03 | − | − | − | − | − | − | − | − | − | + |
| B/Malaysia/2506/04 | − | − | − | − | − | − | + | − | − | − |
| B/Florida/4/06 | − | − | − | − | − | − | − | − | + | + |
| B/Brisbane/3/07 | − | − | − | − | − | − | − | − | + | + |
| B/Bangladesh/3333/07 | − | − | − | − | − | − | − | − | − | + |
| B/England/145/08 | − | − | − | − | − | − | − | − | − | + |
| B/Brisbane/46/08 | − | − | − | − | − | − | − | + | − | − |
| B/Brisbane/60/08 | − | − | − | − | − | − | − | + | − | − |
| B/Hubei‐Wujiagang/158/09 | − | − | − | − | − | − | − | − | − | + |
| B/Wisconsin/1/10 | − | − | − | − | − | − | − | − | − | + |
| Number of reactive strains | 3 | 4 | 7 | 6 | 2 | 1 | 1 | 2 | 4 | 10 |
| Number of ‘forward’ reactive strains | 1 | 4 | 5 | 5 | 2 | 1 | 1 | 2 | 3 | 1 |
NT, not tested: (+): OD ≥ 0·19 (4 standard deviations above the mean background); (−): OD < 0·19 (4 standard deviations above the mean background).
Monoclonal antibodies were raised against the following viruses: A/Brisbane/59/07 (H1N1; A/Bri); A/California/07/09 (H1N1 2009 Pandemic; A/Cal); A/Hiroshima/52/05 (H3N2; A/Hir); A/Singapore/37/09 (H3N2; A/Sin); A/Vietnam/1194/04 (H5N1; A/Vie): A/Indonesia/5/05 (H5N1; A/Ind); B/Malaysia/2506/2004 (B strain; B/Mal); B/Brisbane/60/08 (B strain; B/Bri); B/Panama/45/90 (B strain; B/Pa) and B/Wisconsin/1/10 (B strain; B/Wis). Reactive strains refer to the number of virus isolates that cross‐react with the tested mAb. Forward reactive strains refer to the number of those isolates that cross‐react during or subsequent to the year at which the mAb was raised.
The mAb raised against A/Brisbane/59/07 (H1N1) detected three of eight pre‐pandemic seasonal H1N1 strains and did not detect any of the four pandemic H1N1 influenza viruses tested. The mAb raised against A/California/07/09 (pandemic H1N1) reacted with all four pandemic strains tested, but did not detect any of seasonal H1N1 viruses tested (n = 8).
The mAb raised against A/Hiroshima/52/05 detected all H3N2 strains tested isolated between the years 2000 to 2008 (n = 7), but no H3N2 strains isolated from 1968 to 1999 (n = 15) or post‐2008 (n = 5) were detected. The mAb raised against A/Singapore/37/09 detected all H3N2 isolated post‐2008 strains tested (n = 5) and did not detect H3N2 strains isolated pre‐2009 (n = 8) with the exception of A/Nepal/921/06 (H3N2). The mAb raised against the H5N1 virus A/Indonesia/5/05 (H5N1, clade 2.1) reacted specifically to this virus, whilst the mAb raised against the H5N1 clade 1 virus A/Vietnam/1194/04 detected both strains. The mAb raised against B/Panama/45/90 detected viruses from the B/Yamagata/16/88 lineage isolated between the years 1988 and 2007 (n = 4), but not strains isolated from 2008 onwards (n = 5). The mAb raised against B/Malaysia/2506/04 reacted only against the homologous strain. The mAb raised against B/Brisbane/60/08 reacted with all strains from the B/Victoria/2/87 lineage isolated post‐2007 (n = 2), but not with strains of the B/Yamagata/16/88 lineage. The mAb raised against B/Wisconsin/1/10 reacted with all strains of the B/Yamagata/16/88 lineage isolated between 1994 and 2010 (n = 10), but not with strains from the B/Victoria/2/87 lineage.
These results demonstrate the development of multiple EIAs that can be utilised to detect HA antigen across H1N1, H3N2, H5N1 or B influenza virus strains. Significantly, many of the EIAs exhibited sufficient cross‐reactivity to detect HA antigen from drifted viruses from different seasons. For the mAbs raised against A/California/07/09 (H1N1), A/Hiroshima/52/05 (H3N2), A/Singapore/37/09 (H3N2), A/Vietnam/1194/04 (H5N1) and B/Panama/45/90, this included viruses isolated after the mAb was developed.
EIA shows greater sensitivity than SRID
To measure the sensitivity of EIA, we tested MPH derived from 10 viruses (H1N1, H3N2, H5N1 and B strains) in assays using the relevant mAb to determine the limits of detection (LOD) for the respective EIAs (Table 2). The LOD was calculated using regression analysis to be the HA antigen concentration at which the OD at 450 nm was two standard deviations above the mean background value as described. 13 For EIAs using the homologous mAb (n = 4), the mean sensitivity was 182·87 ng HA/mL (range 26·26–412·85 ng HA/mL). For non‐homologous mAbs (n = 6), a mean sensitivity of 731·62 ng HA/mL (range 12·02–3670·56 ng HA/mL) was observed. These differences in sensitivity were not statistically different (P ≥ 0·05). In comparison, in our laboratory, the LOD for SRID is typically 10 000 ng HA/mL, although it varies between strains (data not shown).
Table 2.
Validation of EIA. Sensitivity, repeatability, intermediate precision and linearity for monovalent pooled harvest (MPH) material (n = 10) by EIA
| MPH strain | mAb | Sensitivity (ng HA/mL) | Repeatability (% CV) | Intermediate precision (% CV) | Linearity (R 2) | |
|---|---|---|---|---|---|---|
| H1N1 | A/Brisbane/59/07 | A/Brisbane/59/07 | 59·39 | 4·8 | 9·4 | 0·993 |
| H3N2 | A/Uruguay/716/07 | A/Hiroshima/52/05 | 78·31 | 1·8 | 4·7 | 0·996 |
| A/Victoria/210/09 | A/Singapore/37/09 | 41·71 | 3·1 | NT | 1 | |
| A/Wisconsin/67/05 | A/Hiroshima/52/05 | 27·27 | NT | NT | NT | |
| A/Brisbane/10/07 | A/Hiroshima/52/05 | 12·02 | NT | NT | NT | |
| H5N1 | A/Indonesia/05/05 | A/Indonesia/5/05 | 232·97 | 4·7 | 8·2 | 0·99 |
| B | B/Brisbane/60/08 | B/Brisbane/60/08 | 412·85 | 3·4 | NT | 0·998 |
| B/Florida/4/06 | B/Panama/45/90 | 3670·56 | 4·2 | 7 | 0·995 | |
| B/Malaysia/2506/04 | B/Malaysia/2506/04 | 26·26 | NT | NT | NT | |
| B/Brisbane/03/07 | B/Panama/45/90 | 559·86 | NT | NT | NT |
NT, not tested; CV, coefficient of variation; HA, haemagglutinin.
The length of time between isolation of the virus used to raise the mAb and the test strain may impact the sensitivity of the EIA. For example, the mAb raised against B/Panama/45/90 was able to detect a virus isolated 16 years later, B/Florida/4/06, albeit with lower sensitivity (3670·56 ng HA/mL).
Intermediate precision and repeatability of EIA compared with SRID
Preparations of MPH derived from six vaccine candidate strains [A/Brisbane/59/07 (H1N1), A/Uruguay/716/07 (H3N2), A/Indonesia/5/05 (H5N1), A/Victoria/210/09 (H3N2), B/Florida/4/06 and B/Brisbane/60/08] were selected to assess repeatability, linearity and intermediate precision of EIA using parameters described in the International Conference on Harmonisation ICH guidelines 17 (Table 2) .
Repeatability was calculated from the coefficient of variation (CV) of samples tested in triplicate by a single operator. The average CV for the six assays was 3·67 ± 1·14% SD (range 1·8–4·8). Linearity was calculated using regression analysis (R 2) of six serial dilutions of MPH material. All assays analysed had an R 2 value of 0·990 or greater. Intermediate precision was calculated to be the percentage CV for virus assayed six times on different occasions by two independent operators, for four of the six strains analysed. The mean % CV was 7·33 ± 2·00% SD (range 4·7–9·4).
Separately, the repeatability and intermediate precision of SRID were calculated for MPH prepared from the strains A/New York/55/04 (H3N2), A/New Caledonia/20/99 (H1N1) and B/Malaysia/2506/04. Repeatability was determined by triplicate analysis of the CV by a single operator for A/New York/55/04. The average CV was 4·57 ± 2·37% SD (range 1·0–8·0). Analysis of the intermediate precision in duplicate, by two operators on two occasions, for the three strains was measured to be 3·75 ± 1·25% SD (range 3–6%) for A/New York/55/04 (n = 8), 7·75 ± 3·55% SD (range 3–12%) for A/New Caledonia/20/99 (n = 8) and 8·25 ± 4·04% SD (range 3–15%) for B/Malaysia/2506/04 (n = 8). Taken together, our results indicate no statistical difference between EIA and SRID in regard to repeatability and intermediate precision when used to determine HA content (P > 0·05 for both repeatability and intermediate precision).
EIA correlates with SRID for measurement of haemagglutinin concentration
The correlation between the EIA and SRID assays for quantitation of HA antigen was determined by testing multiple independent MPH preparations derived from the vaccine candidate strains A/Uruguay/716/07 (H3N2; n = 3), A/Brisbane/10/07 (H3N2; n = 3), A/Victoria/210/09 (H3N2; n = 1), A/Brisbane/59/07 (H1N1; n = 2), B/Brisbane/3/07 (n = 3), B/Florida/4/06 (n = 3) and B/Brisbane/60/08 (n = 1). We demonstrated a linear correlation between the two methods with an R 2 value of 0·957, when all strains were analysed together (Figure 1). Analysis of each strain separately yielded higher levels of correlation: 0·992 for A/Uruguay/716/07, 0·999 for A/Brisbane/10/07, 0·990 for B/Brisbane/3/07 and 0·995 for B/Florida/4/06. These data demonstrate a strong correlation between HA quantitation between EIA and SRID assays.
Figure 1.
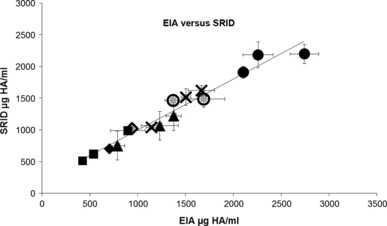
Linearity of HA concentration measured by EIA and SRID Independent monovalent pooled harvest (MPH) preparations (n = 16) were assayed by EIA and SRID, and the concentration of HA antigen plotted against each other. Strains tested include: A/Uruguay/716/07 (cross), A/Brisbane/10/07 (square), A/Brisbane/59/07 (open circle), B/Brisbane/03/07 (triangle), B/Florida/04/06 (circle), A/Victoria/210/09 (open diamond) and B/Brisbane/60/08 (diamond). For EIA analysis of these MPHs, mAbs raised against A/Hiroshima/52/05, A/Hiroshima/52/05, A/Brisbane/59/07, B/Panama/45/90, B/Panama/45/90, A/Singapore/37/09 and B/Brisbane/60/08 were used (respectively). The R 2 was determined to be 0·957. Error bars represent ± 1 SD.
EIA can be used as a stability‐indicating assay
Trivalent vaccine material was held at 2–8°C and sampled at 0, 1, 2, 3 and 6 months. The HA concentration of A/Brisbane/10/07 (H3N2) and B/Brisbane/3/07 was tested using EIA and SRID (Figure 2A,B). Both EIA and SRID assays showed slight fluctuation in HA antigen content over time; however, there was no notable difference between results from the two methods. In separate experiments, vaccine material containing strains A/Wisconsin/67/05 (H3N2) and B/Malaysia/2506/04 was held at RT and sampled on days 0, 1, 4, 7 and 21, and the HA content of these samples was measured by EIA and SRID (Figure 2C,D). Again, no notable differences in the HA antigen content were measured using the two methods.
Figure 2.
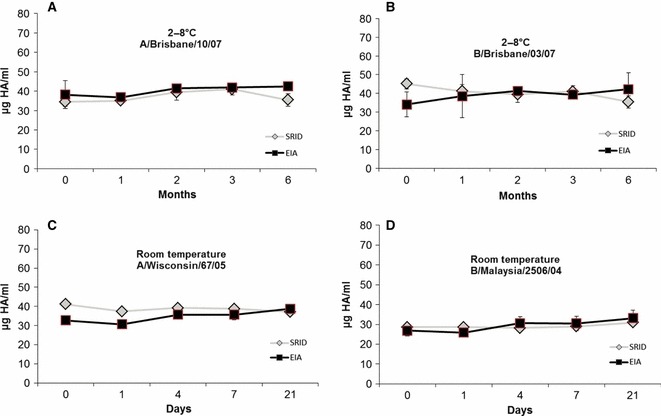
Trivalent vaccine material was held at 2–8°C for 6 months, and the HA concentration was measured by EIA (square) and SRID (diamond) for the strains A/Brisbane/10/07 (A) and B/Brisbane/03/07 (B). The mAbs used were raised against A/Hiroshima/52/05 and B/Panama/45/90, respectively. Trivalent vaccine material was held at room temperature for 21 days, and the HA concentration was measured by EIA (square) and SRID (diamond) for the strains A/Wisconsin/67/05 (C) and B/Malaysia/2506/04 (D). The mAbs used were raised against A/Hiroshima/52/05 and B/Malaysia/2506/04, respectively. Error bars represent ± 1 SD.
The stability‐indicating properties of the EIAs were examined by analysis of trivalent vaccine, which had been subjected to heat (60°C), oxidative (0·001–5% hydrogen peroxide) or pH (pH 2–10) stresses. The stressed vaccines were analysed for the HA content of two strains contained within the vaccine, using both EIA and SRID assays (Figure 3). Generally, the two methods gave similar results. The detection of HA antigen decreased as the heating time or concentration of hydrogen peroxide was increased. When the vaccine preparations were exposed to pH below 5, the content of HA for both viruses was very low or undetected. The HA concentration decreased as the pH increased from 8 to 10, although not as strongly as for low pH. These results demonstrate that the EIA assays described were stability indicating. Whilst the EIAs were consistently more susceptible to the impacts of temperature stress, some examples of SRID having higher susceptibility to hydrogen peroxide and pH stress were observed. Higher than expected fluctuations in the precision of the EIA, especially under chemical stress (Figure 3E,F), were also noted, which were not observed in previous precision studies.
Figure 3.
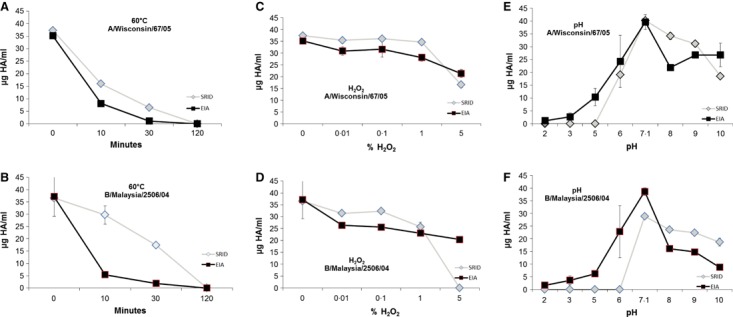
HA concentration of stressed trivalent vaccine material was measured by EIA (square) and SRID (triangle). A/Wisconsin/67/05 (A) and B/Malaysia/2506/04 (B) were held at 60°C for up to 120 minutes. A/Wisconsin/67/05 (C) and B/Malaysia/2506/04 (D) were treated with hydrogen peroxide from 0·001% to 5%. A/Wisconsin/67/05 (E) and B/Malaysia/2506/04 (F) were adjusted pH from pH 2 to 10. For EIA measurement of A/Wisconsin/67/05 and B/Malaysia/2506/04, mAbs raised against A/Hiroshima/52/05 and B/Malaysia/2506/04 were used, respectively. Error bars represent ± 1 SD.
Use of EIA to measure disruption of virus particles for split influenza vaccine
As part of the manufacturing process, it is necessary to analyse the HA content of detergent‐split material from purified virus preparations that have been separated by ultra‐centrifugation over a sucrose gradient. 18 We measured the HA concentration of the different fractions representing different degrees of splitting by EIA and SRID (Figure 4).
Figure 4.
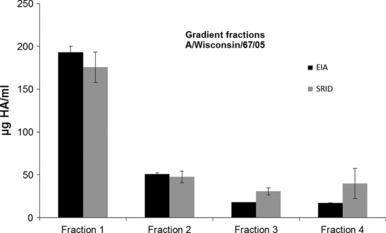
MPH material was separated on a sucrose gradient. Four fractions were collected representing the sedimentation profile for whole and split virus particles for the H3N2 strain A/Wisconsin/67/05. The HA concentration for fractions 1–4 was measured by EIA (black) and SRID (grey). For EIA measurement of A/Wisconsin/67/05, mAb raised against A/Hiroshima/52/05 was used. Error bars represent ± 1 SD.
We observed comparable detection of HA antigen across the four fractions. Whilst some HA content appears slightly higher in fractions 3 and 4 by SRID, this was not statistically significant (P ≥ 0·05; fraction 3 and 4) and may be a consequence of low HA concentration approaching the LOD of the SRID assay for these samples.
EIA can be utilised across multiple years to quantify drifted strains
To understand the impact of EIA on assay development, we assessed the reactivity of drifted strains by EIA. For this purpose, we used historical data for vaccine strain changes that required updating reagents for the SRID assay with each new strain (Table 3). Table 3 outlines the number of strain changes for new strains incorporated into the influenza vaccine from 2000 to 2011, the number of SRID assays developed over this time and the number of EIA assays that could be utilised across these years. From 2000 to 2011, 22 strain changes occurred requiring updates to reagents, and at least two additional reagent sets were developed for H5N1. Thus, 24 sets of reagents were required to be developed and optimised over this 12‐year period. In comparison, our EIA data suggest that only 11 EIA assays would have needed to be developed in order to be able to assess the antigen concentration of drifted strains across this time period. Importantly, several EIAs were effective for quantitating ‘forward’ drifted strains from influenza seasons at or subsequent to the time at which the mAbs were raised. Of the 24 strains active during the 12‐year period, 12 EIAs would have been ‘forward’ reacting and available rapidly following virus drift.
Table 3.
Analysis of influenza vaccine strain changes from 2000 to 2011 including the number of strain changes in this period and the requirements for new antiserum for SRID reagents versus EIA reagents. EIA forward reactive refers to EIA mAbs reactive to strains of the same or subsequent years. Years refer to the number of years within the 12‐year period that EIA reagents would have been available
| Subtype | Strain changes | SRID reagents | EIA reagents | EIA forward reactive | Years |
|---|---|---|---|---|---|
| H1N1 | 4 | 4 | 3 | 2 | 12 |
| H3N2 | 10 | 10 | 3 | 6 | 9 |
| B | 8 | 8 | 4 | 3 | 11 |
| H5N1* | 2 | 2 | 1 | 1 | – |
–: Years available not applicable as was not included in seasonal vaccine.
*Not included in seasonal vaccine.
Discussion
Measurement of HA antigen by an immunological technique is a requirement for registration of influenza vaccine 18 and is essential for the formulation of influenza vaccine potency as well as monitoring product stability for the duration of vaccine administration. Whilst numerous studies have examined the development of alternative assays that are more sensitive, accurate and robust, 6 , 7 , 8 , 10 , 19 SRID continues to be utilised by the majority of vaccine manufacturers to standardise influenza vaccine. This is because it satisfies requirements for an immunological assay for HA as recommended by a World Health Organisation Expert committee 8 and is a listed requirement in the British Pharmacopoeia. 18
In this report, we demonstrated EIA was comparable to the SRID assays with respect to measuring levels of influenza antigen and also demonstrated improved sensitivity and comparable precision. The LOD for SRID (approximately 10 000 ng HA/mL) was significantly higher than any of the EIA assays described here. For EIA, the mAbs used were selected by their HA‐specific HAI ability, suggesting reactivity to the receptor binding site of HA. 20 This site induces high levels of neutralising antibodies upon vaccination. 3 , 21 We suggest that utilising the same mAb raised against a unique epitope site for capture and detection in a sandwich EIA limits detection to native, trimeric (and higher order complexes of) HA. Denatured and monomeric HA molecules are unlikely to be detected. Significantly, this corresponds to detection of the HA species (native, trimer) considered optimal for eliciting neutralising antibodies 21 and thus most effective for vaccine material. Supporting this, mice vaccinated with physically or chemically stressed vaccine produced significantly lower levels of HA‐neutralising antibodies and were less protected against a lethal influenza challenge. 22 , 23
We have demonstrated decreased detection of HA upon chemical and physical stresses suggesting EIA, like SRID, can measure vaccine stability. The EIA detection of temperature‐stressed HA was consistently lower for both influenza A and B viruses compared with SRID, suggesting EIA may have greater sensitivity for detecting temperature impacts on vaccine material. Further, Epand and co‐workers indicated that heat stress of a H1N1 and H3N2 virus between 55 and 65°C resulted in a decrease of trimeric HA present and an increase in monomeric HA present, 24 providing further evidence that EIA only detects trimeric, native HA. Assessing the stability of HA in response to chemical stresses was less clear. Chemical stresses may have had more subtle impacts on protein structure and folding, thus differing the impacts on binding of mAb and polyclonal sera. Whilst SRID was more sensitive to chemical stress than EIA in some instances, a more detailed study would be required to understand the differences in detection. Fluctuations in the precision of EIA were also observed for HA subjected to a pH of 6, for both A/Wisconsin/67/05 (H3N2) and B/Malaysia/2506/04 at a higher rate to that of SRID. This was not observed during other precision analysis and may be because of structural instability of stressed, trimeric, HA. This observation may not have been apparent in material tested by SRID, which also detects HA in non‐native and monomeric forms, as well as possible non‐specific binding, because of the use of polyclonal antiserum.
Our data demonstrated that a single EIA often cross‐reacted with drifted strains over several years. As an example, the mAb raised against B/Wisconsin/1/10 cross‐reacted with viruses of the B/Yamagata/16/88 lineage between 1994 and 2000, a period of 17 years. We also demonstrated that several EIAs were effective for quantitating ‘forward’ drifted stains isolated in or subsequent to the year at which the mAb was raised. Over a 12‐year period examined, this corresponded to 12 instances where EIA mAb reagents would have been rapidly available following virus drift. In contrast, SRID reagents have limited utility for drifted strains with the development of new reagents highly recommended for each new candidate strain. 19 There are reports of cross‐reactive polyclonal antiserum being utilised in SRID against newly drifted strains 25 ; however, to our knowledge, the use of cross‐reactive SRID reagents to drifted strains over a period of several years has not been extensively published. We suggest that EIA has the potential to reduce the number of reference reagents that need to be developed when compared with SRID.
The generation of polyclonal sera used in SRID takes approximately 12–16 weeks to produce and is reliant on obtaining purified HA antigen. The purification of HA may be impacted by susceptibility to enzyme degradation and has previously delayed the development of SRID reagents and consequently release of vaccine. 26 In contrast, generation of mAb for EIA does not require purification of HA antigen and, in our laboratory, it takes approximately 6 weeks to reach the validation stage of the EIA.
The generation of large quantities of reference antigen is required for both EIA and SRID assays. This represents a bottleneck in the development of assay reagents that has the potential to delay the release of EIA for quantitation of HA antigen. Strategies to rapidly standardise reference antigen for use in the EIA are currently being investigated as well as using existing reference antigen to accurately determine potency of drifted strains.
We suggest that due to its increased sensitivity, detection of drifted strains, reduction in development time and the possibility of automation, the EIA assay provides a robust alternative to the SRID assay that is currently used for the quantitation of HA antigen in influenza virus vaccine. The simplicity of this EIA would also allow for rapid transfer to laboratories around the world.
Conflicts of interest
The WHO Collaborating Centre for Reference and Research on Influenza has received funding from CSL Limited for performing serology and other contract research work that was not related to this current publication. IB, RS, JB and SR own a small number of shares in CSL Limited. JB, SR, CO, EV and KV are employees of CSL Limited.
References
- 1. WHO . Influenza (seasonal) fact sheet. 2009. Available at: http://www.who.int/mediacentre/factsheets/fs211/en/index.html (Accessed 12th November 2009).
- 2. Wiley DC, Wilson IA, Skehel JJ. Structural identification of the antibody‐binding sites of Hong Kong influenza haemagglutinin and their involvement in antigenic variation. Nature 1981; 29:289. [DOI] [PubMed] [Google Scholar]
- 3. Knossow M, Gaudier M, Douglas A et al. Mechanism of neutralization of influenza virus infectivity by antibodies. Virology 2002; 302:294–298. [DOI] [PubMed] [Google Scholar]
- 4. The Department of Health GB . British Pharmacopoeia: Crown Copyright. 2003.
- 5. Wood JM, Schild GC, Newman RW, Seagroatt V. Application of an improved single‐radial‐immunodiffusion technique for the assay of haemagglutinin antigen content of whole virus and subunit influenza vaccines. Dev Biol Stand 1977; 39:193–200. [PubMed] [Google Scholar]
- 6. He Q, Velumani S, Du Q et al. Detection of H5 avian influenza viruses by antigen‐capture enzyme‐linked immunosorbent assay using H5‐specific monoclonal antibody. Clin Vaccine Immunol 2007; 14:617–623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kapteyn JC, Saidi MD, Dijkstra R et al. Haemagglutinin quantification and identification of influenza A&B strains propagated in PER.C6 cells: a novel RP‐HPLC method. Vaccine 2006; 24:3137–3144. [DOI] [PubMed] [Google Scholar]
- 8. Mayner RE, Blackburn RJ, Barry DW. Quantitation of influenza vaccine hemagglutinin by immunoelectrophoresis. Dev Biol Stand 1977; 39:169–178. [PubMed] [Google Scholar]
- 9. van de Donk HJ, de Jong JC, van Olderen MF, Osterhaus AD. Monoclonal antibodies for the control of influenza virusvaccines. Dev Biol Stand 1984; 57:251–255. [PubMed] [Google Scholar]
- 10. Williams TL, Luna L, Guo Z et al. Quantification of influenza virus hemagglutinins in complex mixtures using isotope dilution tandem mass spectrometry. Vaccine 2008; 26:2510–2520. [DOI] [PubMed] [Google Scholar]
- 11. Pietrzykowski E, Margetts MB, MacFarlan RI, Webb EA, Cox JC, Edwards SJ. Antibody responses to HPV6b E polyproteins and production of monoclonal antibodies. Hybrid Hybridomics 2002; 21:333–338. [DOI] [PubMed] [Google Scholar]
- 12. Dowdele W, Kendal A, Noble G. Influenza viruses; in Lenette E, Schmidt N. (eds): Diagnostic Procedure for Viral, Rickettsial and Chlamydial Infections, 5 edn Washington, DC: American Public Health Association, 1979; 585–609. [Google Scholar]
- 13. Bhatti A, Siddiqui YM, Micusan VV. Highly sensitive fluorogenic enzyme‐linked immunosorbant assay: detection of staphylococcal enterotoxin B1. J Microbiol Methods 1994; 19:179–187. [Google Scholar]
- 14. Williams MS, Mayner RE, Daniel NJ et al. New developments in the measurement of the hemagglutinin content of influenza virus vaccines by single‐radial‐immunodiffusion. J Biol Stand 1980; 8:289–296. [DOI] [PubMed] [Google Scholar]
- 15. Brownlee K. Statistical Theory and Methodology in Science and Engineering. New York: John Wiley and Sons, 1965. [Google Scholar]
- 16. Sokal RR, Rohlf FJ (eds). Biometry: The Principles and Practice of Statistics in Biological Research. New York: Freeman, 1994. [Google Scholar]
- 17. ICH Harmonised Tripartite Guidlines: Validation of Analytical Procedures: Text and Methodology Q2(R1). Available at: http://www.ich.org/products/guidelines/quality/article/quality‐guidelines.html (Accessed 1 February 2010).
- 18. British Pharmacopoeia 2009 . Influenza Vaccine (Split Virion Inactivated) Incorporating the Requirements of the European Pharmacopoeia 2002. Publisher: TSO (the stationary Office), Norwich, Norfolk, UK: Crown Copyright, 2009; 3166–3169. [Google Scholar]
- 19. Schild GC, Wood JM, Newman RW. A single‐radial‐immunodiffusion technique for the assay of influenza haemagglutinin antigen. Proposals for an assay method for the haemagglutinin content of influenza vaccines. Bull World Health Organ 1975; 52:223–231. [PMC free article] [PubMed] [Google Scholar]
- 20. Weis W, Brown JH, Cusack S, Paulson JC, Skehel JJ, Wiley DC. Structure of the influenza virus haemagglutinin complexed with its receptor, sialic acid. Nature 1988; 333:426–431. [DOI] [PubMed] [Google Scholar]
- 21. Wei CJ, Xu L, Kong WP et al. Comparative efficacy of neutralizing antibodies elicited by recombinant hemagglutinin proteins from avian H5N1 influenza virus. J Virol 2008; 82:6200–6208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Quan FS, Li ZN, Kim MC et al. Immunogenicity of low‐pH treated whole viral influenza vaccine. Virology 2011; 417:196–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Quan FS, Huang C, Compans RW, Kang SM. Virus‐like particle vaccine induces protective immunity against homologous and hetrologous strains of influenza virus. J Virol 2007; 81:3514–3524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Epand RM, Epand RF. Thermal denaturation of influenza virus and its relationship to membrane fusion. Biochem J 2002; 365:841–848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Vodeiko GM, Weir JP. Determination of H5N1 vaccine potecy using reference antisera from hetrologous strains of influenza. Influenza Other Respir Viruses 2011; 6, 176–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Rockman S, Schoofs P, Greenburg M. Development and testing of the Australian pandemic influenza vaccine – a timely response. Microbiol Aust 2011; 32:10–14. [Google Scholar]


