Abstract
Ionotropic glutamate receptors (iGluRs) are ligand-gated ion channels that mediate the majority of excitatory neurotransmission in the central nervous system. iGluRs open their ion channels in response to binding of the neurotransmitter glutamate, rapidly depolarize the postsynaptic neuronal membrane, and initiate signal transduction. Recent studies using X-ray crystallography and cryo-electron microscopy have determined full-length iGluR structures that (1) uncover the receptor architecture in an unliganded, resting state, (2) reveal conformational changes produced by ligands in order to activate iGluRs, open their ion channels, and conduct ions, and (3) show how activated, glutamate-bound iGluRs can adopt a nonconducting desensitized state. These new findings, combined with the results of previous structural and functional experiments, kinetic and molecular modeling, mutagenesis, and biochemical analyses, provide new views on the structural mechanisms of iGluR gating.
Ionotropic glutamate receptors (iGluRs) are ligand-gated ion channels that mediate excitatory neurotransmission throughout the central nervous system (CNS).1 Aberrancies in iGluR function result in a wide range of neurological diseases.1−4 Glutamate, the primary neurotransmitter at almost all synapses in the CNS, is released from presynaptic terminals and binds to postsynaptic iGluRs, which in response open their ion channels for the flow of cations, rapidly depolarize the postsynaptic membrane, and initiate signal transduction in the postsynaptic neuron. The iGluR family of integral membrane proteins includes four subtypes in vertebrates: α-amino-3-hydroxy-5-methyl-4-isoaxazolepropionic acid (AMPA), kainate (KA), N-methyl-d-aspartate (NMDA), and δ-receptors. Each iGluR family member exhibits specific kinetic and pharmacological properties, in addition to playing unique roles in neurotransmission.
Despite their diversity, iGluRs share a common modular structural design (Figure 1).5,6 All iGluRs are assemblies of four similar or identical subunits (A–D) organized into a layered architecture. A large extracellular domain (ECD) comprises the amino-terminal domain (ATD) layer that is necessary for assembly, trafficking, and functional regulation of the receptor1,7 and the ligand binding domain (LBD)8 layer, which sits below the ATD and harbors binding sites for ligands that activate, modulate, or antagonize the receptor. Below the ATD and LBD layers is the transmembrane domain (TMD)9 that forms a channel for cations to flow through the postsynaptic membrane. The cytoplasmic carboxy-terminal domains (CTDs), which are involved in receptor localization and regulation,10 have widely different sizes depending on the iGluR subunit. These likely form an intracellular fourth layer but have so far evaded structural determination.
Figure 1.
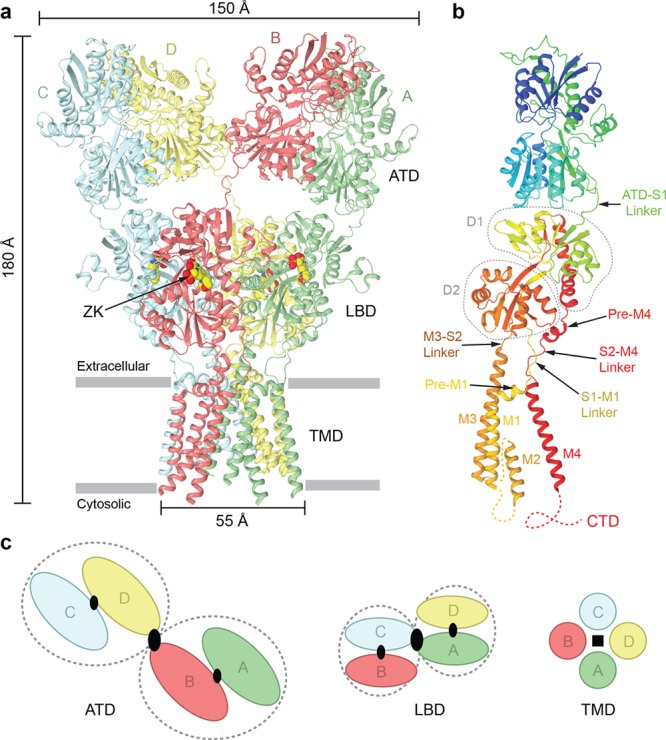
iGluR structural architecture and domain arrangement. (a) Crystal structure of a homotetrameric AMPAR composed of GluA2 subunits in the closed, competitive antagonist ZK200775-bound state [Protein Data Bank (PDB) entry 3KG2] viewed parallel to the membrane. Each of the four GluA2 subunits is colored differently: green (A), red (B), blue (C), and yellow (D). ZK200775 molecules are shown as space-filling models. (b) Single GluA2 subunit (A/C), rainbow-colored from blue (N-terminus) to red (C-terminus). The LBD upper (D1) and lower (D2) lobes are indicated by gray, dashed contours. (c) Representations of each iGluR domain layer (ATD, LBD, and TMD) viewed extracellularly, parallel to the axis of the receptor overall 2-fold rotational symmetry. The axes of local 2-fold symmetry in the ATD and LBD dimers are indicated with small black ovals and those of overall 2-fold symmetry with large black ovals, and local 4-fold symmetry in the TMD is indicated with a black square. Gray, dashed contours encapsulate local dimer pairs in the ATD and LBD layers.
Overall, the structures of iGluRs are 2-fold symmetric, unique for tetrameric ion channels, where cross-subunit interactions and symmetry partners swap along the three-layered topology of the receptors11 (Figure 1c). In the ATD layer, local dimers between subunits A and B and subunits C and D form on each side of the overall 2-fold symmetry axis, with a cross-dimer interface between subunits B and D. In the LBD layer, the local dimer pairs are switched, subunits A and D and subunits B and C, and the cross-dimer interface is formed between subunits A and C. Each LDB is comprised of two polypeptide stretches, S1 and S2, which form a clamshell structure, with ligand binding occurring in the middle, between the LDB upper (D1) and lower (D2) lobes (Figure 1b). When the agonist binds, the LBD clamshells close to induce channel opening in the TMD. The TMD, which has an inverted orientation in the membrane compared to voltage-gated ion channels, consists of three transmembrane helices (M1, M3, and M4) and a re-entrant intracellular loop M2 between helices M1 and M3. The LBDs are tethered to the TMD through the flexible polypeptide linkers S1–M1, M3–S2, and S2–M4. These linkers communicate conformational changes in the LBD induced by binding of the ligand to the TMD. While the linker region is still 2-fold symmetric, the membrane-residing TMD is nearly 4-fold symmetric. The M3 segments line the extracellular portion of the ion channel pore, while M1 and M4 surround M3s and form the ion channel periphery. The re-entrant loop M2 includes an N-terminal helix, which forms extensive cross-subunit interfaces, and an extended C-terminal region, which lines the intracellular portion of the ion channel pore and forms a selectivity filter.
While the structures of full-length receptors have been well-studied by X-ray crystallography11−17 and cryo-electron microscopy (cryo-EM),13,18−24 the process by which iGluRs bind glutamate to conduct cations into the postsynaptic density has remained elusive. Only recently have cryo-EM studies succeeded in determining the first high-resolution structure of an activated, glutamate-bound AMPA receptor (AMPAR) in a conducting state25 and also in visualizing the conformational changes in the entire receptor during desensitization.26 Coupled with earlier X-ray crystallographic studies of full-length AMPARs in unliganded13,17 (apo) and pre-active states,12,13,16 we can begin to visualize the entire gating mechanism of iGluRs using AMPARs as a template. This Perspective outlines recent progress in AMPAR structural biology to provide an in-depth visualization of the complete iGluR gating mechanism for the first time.
Gating in AMPARs
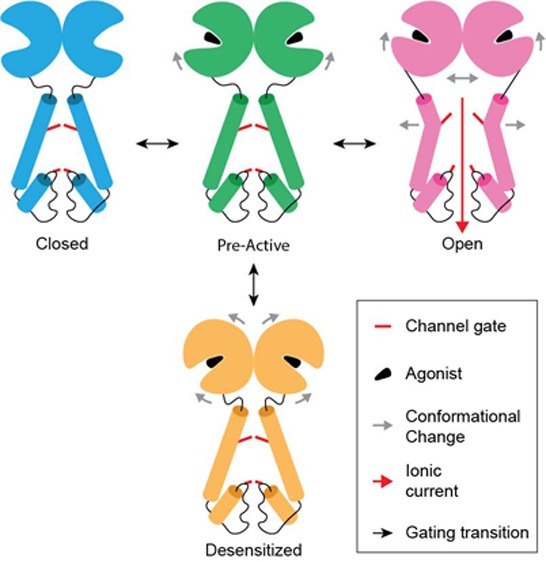
We refer to gating in iGluRs, and in this specific case of AMPARs, as the general series of conformational changes in the receptor that occur upon ligand binding or unbinding to open or close the ion channel and affect the functional state of the receptor. A whole-cell patch-clamp current in response to a prolonged application of agonist glutamate (Figure 2a) illustrates three major iGluR gating functions: activation, desensitization, and deactivation. iGluR gating functions can also be described by a simplified kinetic model that includes closed, pre-active, open, and desensitized states (Figure 2b). In the absence of agonist (A), the receptors reside in a resting, nonconducting state C (closed). The concentration-dependent process of agonist binding (C to CA transition) is immediately followed by conformational changes that place the receptor in a pre-active state (P), where it is ready to follow one of the two major functional routes: the receptor can either convert into a conducting (indicated by an asterisk) state O (open) or adopt an active but nonconducting state D (desensitized). The PA to OA transition is much faster than the PA to DA transition and defines the fast, submillisecond time scale rise in the inward current signifying the activation process (Figure 2a). Typically, however, the equilibrium between agonist-bound states is strongly shifted toward DA, and the majority of the receptors (95–99% for AMPARs) undergo the transition from OA to DA, underlying the current decline phase in the continuous presence of agonist and signifying the desensitization gating process (Figure 2a). Note that there is no direct transition between OA and DA27 and receptors have to visit PA between those states. After agonist removal, receptors undergo the transition from all agonist-bound states (the majority are in DA) to the closed state C defining the recovery of the steady state current to zero in the process of deactivation (Figure 2a).
Figure 2.
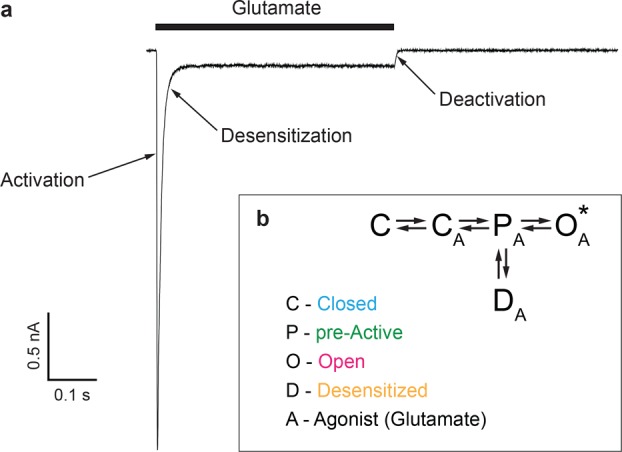
Functional recording and kinetic model of iGluR gating. (a) Representative whole-cell patch-clamp current recorded at a −60 mV membrane potential from a HEK-293 cell expressing GluA2 in response to a 0.5 s application of the agonist glutamate (black bar). Current components that are mainly determined by the three major gating processes, activation, desensitization, and deactivation, are indicated by arrows. (b) Simplified kinetic model of iGluR gating, with closed (C), pre-active (P), open (O), and desensitized (D) states. The agonist-bound states are indicated by “A”, and the conducting state is indicated by an asterisk. Colors associated with each state C (blue), P (green), O (magenta), and D (orange) are used throughout this Perspective for structural representations.
Despite the fact that the basic gating processes can be described by the simplified kinetic model (Figure 2b), the detailed analysis of iGluR activity typically requires more complicated kinetic models.16,27−30 For example, AMPAR single-channel currents show multiple conductance levels,31,32 and occupancy of these levels depends on the agonist type and concentration,30,33−37 reflecting different extents of pore opening as well as the activation state and coupling efficiency for each of the four contributing receptor subunits. While kinetic models can account for these multiple conductance levels by introducing additional subunit-specific states, the structural bases underlying transitions between these states remain obscure. We will therefore discuss how the kinetic transitions occur at the structural level in the framework of the simplified kinetic model, based on cryo-EM and X-ray crystallography structures of full-length AMPARs.
Structures Describing the AMPAR Gating States
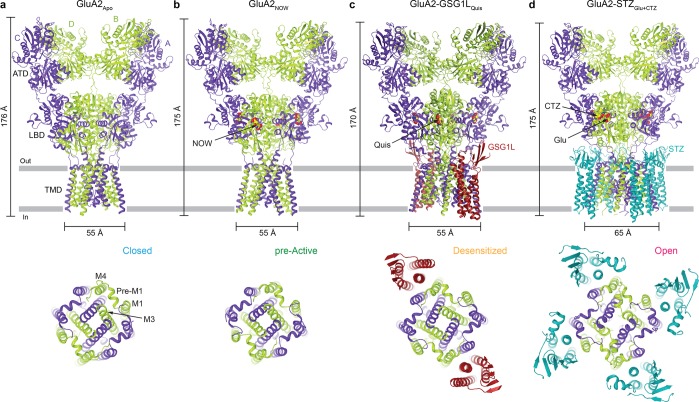
Since the first structure of a full-length iGluR11 (AMPAR subtype GluA2) in the closed, competitive antagonist ZK200775 (ZK)-bound state was determined, significant progress has been made by both crystallographic and cryo-EM studies, which revealed the key states in the AMPAR gating mechanism (Figure 3). For example, two available apo state GluA2 crystal structures13,17 can be used to describe closed state C in the gating model. Figure 3a illustrates one of the two, GluA2Apo,17 with a more symmetrical arrangement of domains and a better-resolved TMD. Also determined by crystallography, there is a range of agonist-bound structures,12,13,16 with partially closed LBD clamshells bound to agonist and closed ion channels. Although it is yet unclear how functionally identified pre-active states relate to these structures, in the first approximation they can serve as models of PA. In this Perspective, we use the crystal structure of GluA2 in complex with the partial agonist nitrowillardiine (NOW)16 (GluA2NOW) as a representative structure for the pre-active state PA (Figure 3b).
Figure 3.
Structures of AMPARs and AMPAR complexes representing different gating conformations. Shown are the entire structures viewed parallel to the membrane (top row) or their TMDs viewed extracellularly (bottom row). (a) Crystal structure of GluA2 in the absence of ligands, GluA2Apo (PDB entry 5L1B), representing the resting, closed state (C). (b) Crystal structure of GluA2 bound to the partial agonist nitrowillardiine (NOW), GluA2NOW (PDB entry 4U4F), representing the pre-active state (P). (c) Cryo-EM structure of the GluA2–GSG1L complex bound to the full agonist quisqualate (Quis), GluA2–GSG1LQuis (PDB entry 5VHZ), representing the desensitized state (D). (d) Cryo-EM structure of the GluA2–STZ complex bound to the full agonist glutamate (Glu) and positive allosteric modulator cyclothiazide (CTZ), GluA2–STZGlu+CTZ (PDB entry 5WEO), representing the open state (O). All ligands are shown as space-filling models. AMPAR subunits are colored purple (A and C) and green (B and D), with auxiliary subunits colored red (GSG1L) or blue (STZ).
Additional structures of AMPARs that we use as structural representations of states in the AMPAR gating mechanism were determined using cryo-EM in the presence of transmembrane auxiliary subunits. These auxiliary subunits assemble with the majority of AMPARs in vivo and alter their kinetics and pharmacology to specifically modulate receptor function at various synapses throughout the CNS. Several comprehensive reviews describe these regulatory subunits in great detail.38−41 One such regulatory subunit, germline-specific gene1-like (GSG1L),42,43 reduces the AMPAR channel open probability and favors nonconducting states of the receptor.44,45 Cryo-EM was used to determine the structure of the GluA2–GSG1L complex in the presence of the antagonist ZK25 (GluA2–GSG1LZK) to an overall resolution of 4.6 Å, with a higher local resolution in the TMD (∼4 Å) to provide the most complete closed state iGluR channel structure to date. As this structure is nearly identical to that of GluA2Apo yet has better-resolved molecular details in the TMD, we use GluA2–GSG1LZK to describe closed state C in the gating mechanism throughout the remainder of this Perspective.
Because GSG1L also favors AMPAR desensitization, we determined the first AMPAR structure in the desensitized state26 by applying cryo-EM to the GluA2–GSG1L complex bound to the high-affinity agonist quisqualate (Quis), which slows down recovery from desensitization compared to the lower-affinity agonist glutamate.46 We use this GluA2–GSG1LQuis structure (Figure 3c) to represent the desensitized state DA in the AMPAR gating model (Figure 2b). Contrary to GSG1L, the auxiliary subunit stargazin (STZ) increases the channel open probability and favors the AMPAR open state.37,47 Correspondingly, we applied cryo-EM to the GluA2–STZ complex bound to the agonist glutamate and the positive allosteric modulator cyclothiazide (CTZ) to determine the first high-resolution (3–4 Å local resolution throughout the TMD) structure of an activated iGluR with the ion channel in the open state.25 We use this GluA2–STZGlu+CTZ structure (Figure 3d) to represent the open state OA in the AMPAR gating model.
We assume that the structures describing the gating states of AMPARs in the absence or presence of auxiliary subunits are largely similar. This assumption is strongly supported by nearly identical structures of AMPARs in the apo or antagonist-bound states in the absence11−13,16,20,48 or presence20,21,25,26 of auxiliary subunits. However, such a comparison is not yet available for other gating states. In addition, all currently available structures of AMPAR receptor complexes are with claudin-like auxiliary subunits,20,21,25,26,49 while other types of auxiliary subunits may exert stronger influences on AMPAR conformations. We also model gating transitions by morphing the discrete gating states. Perhaps intermediate gating states, which are currently unavailable, and unapproachable by traditional X-ray crystallography and cryo-EM methods, will be resolved in the future using nuclear magnetic resonance spectroscopy, X-ray free electron lasers, or time-resolved cryo-EM.
LBD: The Gating Initiation Domain
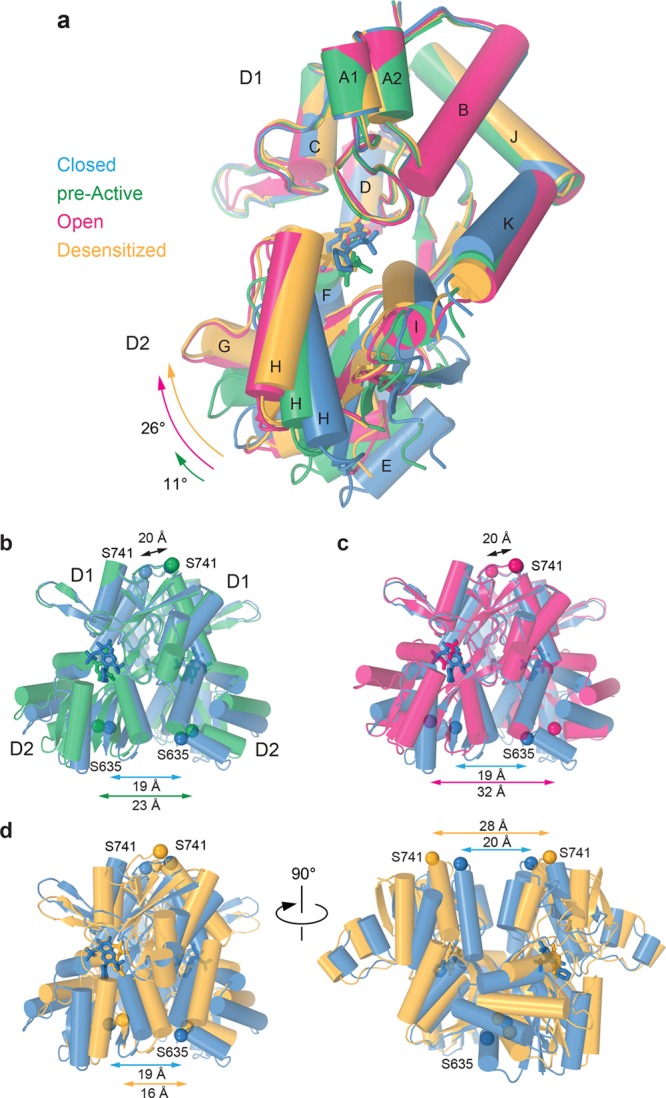
LBDs are the chemical recognition sites in AMPARs, acting as gating initiation domains to communicate the presence or absence of ligands to the rest of the receptor and TMD in particular. In the resting or closed state, C, the clamshell of an individual LBD (Figure 4a and Movie 1) is maximally open (true for both the apo and antagonist-bound states). When the agonist binds, the clamshell closes, with D2 moving toward D1, reducing the size of the cleft between D1 and D2.50 Full clamshell closure, resulting in the maximally open (OA) or desensitized (DA) states, is characterized by a 26° swing of D2 toward D1. Intermediate clamshell closure (11° in the GluA2NOW structure) characterizes the pre-active state, PA, where the channel remains completely closed and no conformational changes associated with opening or desensitization have yet happened (see below). Recent biophysical studies suggested that binding of any given agonist can induce different extents of LBD clamshell closure, but the extent of iGluR activation depends on the probability of the LBD clamshell to occupy its completely closed conformation.30,51−56 Accordingly, compared to full agonists, partial agonists less frequently elicit complete closure of the LBD clamshell and induce channel opening with lower probability. Consistent with this idea, incompletely closed clamshells are easier to capture in structures with partial agonists, especially if these structures are stabilized by crystal contacts or complexes with auxiliary subunits.13,16,49
Figure 4.
Structural rearrangements in individual LBDs and LBD dimers during gating. (a) Superposition of LBD monomers from GluA2–GSG1LZK-1 (blue, PDB entry 5WEK), GluA2NOW (green, PDB entry 4U4F), GluA2–GSG1LQuis (orange, PDB entry 5VHZ), and GluA2–STZGlu+CTZ (magenta, PDB entry 5WEO) structures based on the upper lobe D1. Movement of the lower lobe D2 relative to the GluA2–GSG1LZK-1 structure is illustrated with arrows of the corresponding color. (b–d) Superposition of LBD dimers from GluA2–GSG1LZK-1 and (b) GluA2NOW, (c) GluA2–GSG1LQuis, or (d) GluA2–STZGlu+CTZ. Cα atoms of S635 and S741 are shown as spheres of the corresponding color with cross-dimer distances between them indicated.
At the level of LBD dimers (Figure 4b–d and Movie 2), it becomes apparent how the same extent of closure of the individual LBD clamshells can result in the completely different gating functions of activation and desensitization.50,57,58 We use distances between Cα atoms of S741 and S635 to measure changes in the LBD dimer D1–D1 and D2–D2 lobe separation, respectively, in different gating states (Figure 4b–d). Because of the back-to-back dimer arrangement, maintaining the D1–D1 interface during activation allows conversion of the individual clamshell closures to dramatic separation of the D2 lobes (Figure 4c). It is this separation that is used to open the iGluR ion channel. Alternatively, the D1 lobes of the LBD dimers become separated from each other during desensitization as a result of back-to-back rolling of the individual LBDs. Because the individual LBD clamshells remain maximally closed, separation of the D1 lobes brings the D2 lobes closer together (Figure 4d), releasing strain on the linkers leading to the ion channel and thus leaving it in a nonconducting state. However, before the LBD clamshells proceed with one of these two dramatically different conformational pathways, they first reach a pre-active conformation with partially closed clamshells (Figure 4b). In essence, PA is a bifurcation point in the iGluR gating energy landscape: here the energy of agonist binding goes into partial closure of the individual LBD clamshells, creating strain on both the D1–D1 interface and the linkers connecting D2s to the ion channel; this energy can further be used to either break the M3 bundle seal and open the ion channel for conductance or rupture the D1–D1 interface to keep the D2 lobes close together and not disturb the closed channel conformation.
The changes in the LBD local dimers happen 2-fold symmetrically across the overall 2-fold rotational symmetry axis of the receptor, and this paired movement is what acts to change the gating state of the TMD below (Figure 5 and Movie 3). In the resting, closed state (C), the LBDs create a tight arrangement around the central receptor axis; the state of the ion channel is directly affected by movement of LBDs away from this axis to strain the LBD–TMD linkers. Only a little gross difference in the overall LBD layer arrangement is observed between C and PA (Figure 5a). Much more significantly, the LBD layer expands in OA, where the separation of the D2 lobes in local LBD dimers results in an overall movement of the LBDs away from the 2-fold symmetry axis (Figure 5b), which ultimately adds tension on the LBD–TMD linkers to open the ion channel. In the desensitized state, DA, regardless of maximum clamshell closure in individual LBDs, the ruptured D1–D1 LBD dimer interfaces allow subunits A and C LBDs to rotate 14° away from their dimer pair partners (Figure 5c). These structural rearrangements result in the loss of the local 2-fold rotational symmetry in the LBD dimers, make the LBD layer appear more 4-fold symmetrical, and bring the individual LBDs closer to the axis of the overall 2-fold symmetry (Figure 5c), thus allowing the ion channel to stay closed.
Figure 5.
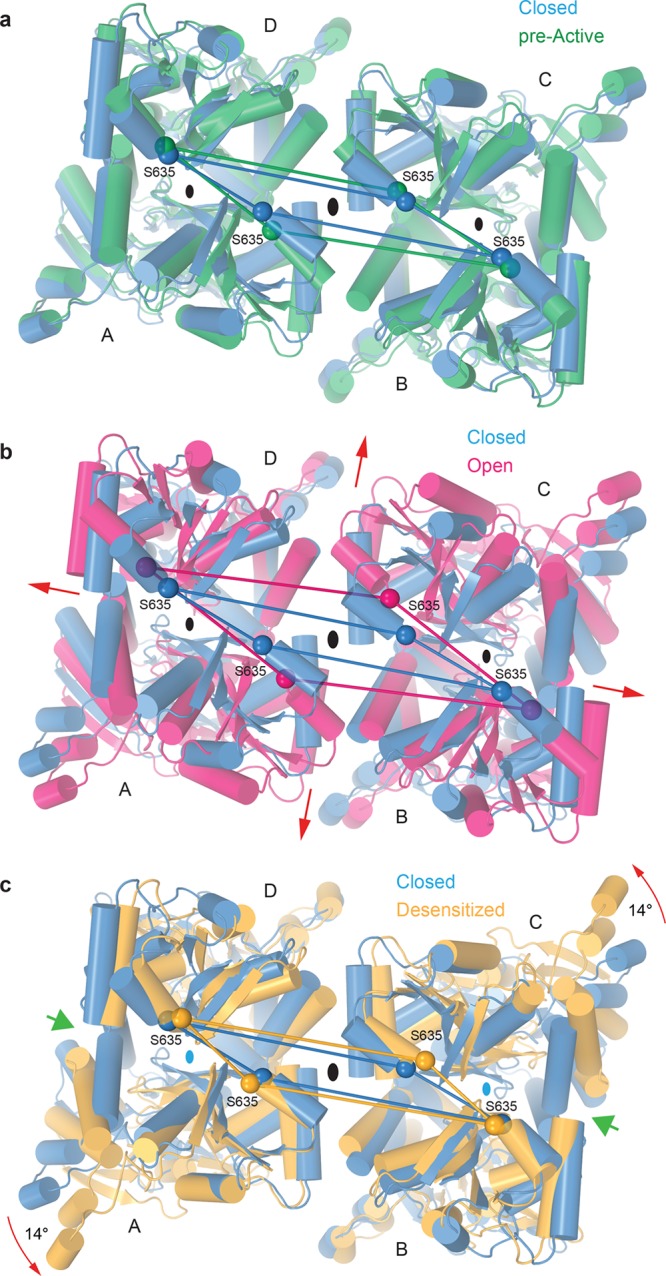
Structural rearrangements in the LBD tetramer during gating. Superposition of LBD tetramers from GluA2–GSG1LZK-1 (blue, closed state) and (a) GluA2NOW (green, pre-active state), (b) GluA2–STZGlu+CTZ (magenta, open state), or (c) GluA2–GSG1LQuis (orange, desensitized state) viewed from the TMD along the axis of the overall 2-fold receptor symmetry (large black ovals in the middle). Cα atoms of S635 are shown as spheres of the corresponding color, connected by straight lines. Broadening of the LBD layer in the open state and rotation of the A and C monomers in the desensitized state are indicated by red arrows. Green arrows in panel c point to the cleft between the desensitized state LBD protomers, signifying the loss of local 2-fold symmetry in LBD dimers (small ovals) and 3-fold smaller intradimer interfaces.
LBD–TMD Linkers: The Gating Transmission Elements
The aforementioned changes in the LBD layer are transmitted to the ion channel by means of the LBD–TMD linkers. Because of the 2-fold symmetrical arrangement of the LBDs, the linkers connecting them to the pseudo-4-fold symmetrical TMD form two conformationally distinct diagonal pairs that play different roles in the geometric and energetic coupling of protein domains in subunits A and C versus subunits B and D and correspondingly define the distinct roles of the two pairs of diagonal subunits in iGluR gating. During gating, the three pairs of LBD–TMD linkers, S1–M1, M3–S2, and S2–M4, in the A and C as well as B and D diagonal pairs undergo significant conformational changes (Movie 4, Movie 5, and Figure 6). The principal changes that drive the ion channel to open or close occur in the B and D subunit M3–S2 linkers, which splay apart by 12 Å to pull on the M3 pore-forming helices (Figure 6b). The same linkers in the A and C subunits undergo little change (Figure 6a) and move closer together by 1 Å. These observations are in good agreement with studies showing that the B and D subunits play a more important and direct role in gating than the A and C subunits do.59,60 Similarly, the S1–M1 linkers in the B and D subunits splay apart by 13 Å, while the linkers in the A and C subunits separate by only 5 Å. In the last set of LBD–TMD linkers, the S2–M4 linkers, the B and D subunits maintain their relative positioning. Surprisingly, drastic changes are observed in the A and C linkers, which include complete unwinding of the pre-M4 helices and stretching of the S2–M4 linkers toward the central pore axis to contribute to the ion permeation pathway (Figure 6c,d and Movie 4). This finding helps to explain why the M4 segments are critical for iGluR tetrameric functional assembly61−63 and why mutations in this region are related to human pathologies and have dramatic effects on iGluR gating kinetics.64−66
Figure 6.
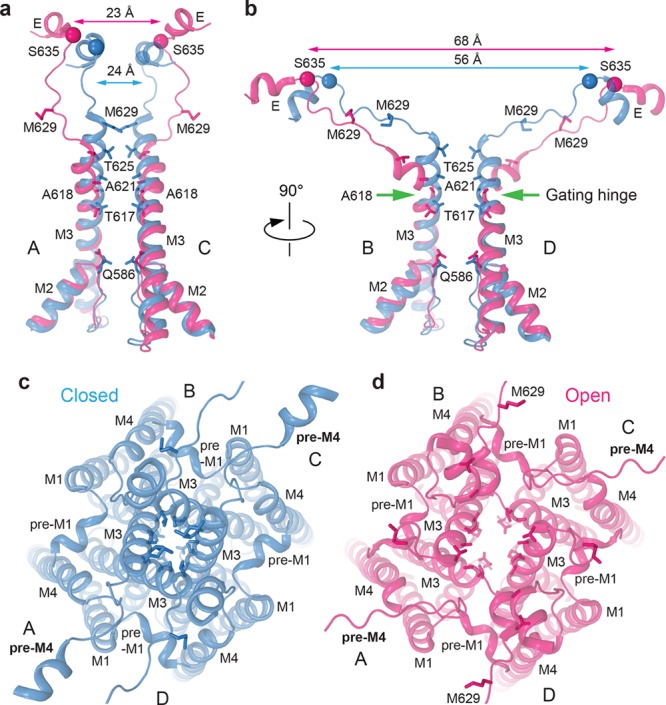
Structural rearrangements in the ion channel during opening. (a and b) Close-ups of the superposition of pore-lining domains M2 and M3 and M3–S2 linkers in subunits (a) A and C and (b) B and D of GluA2–GSG1LZK-1 (blue, closed state) and GluA2–STZGlu+CTZ (magenta, open state). Cα atoms of S635 are shown as spheres of the corresponding color with cross-dimer distances between them indicated. Residues forming the upper (T617, A621, T625, and M629) and lower (Q586) gates as well as the gating hinge alanine A618 are shown as sticks. The location of the gating hinge in subunits B and D is indicated by green arrows. Extracellular view of the ion channel in (c) GluA2–GSG1LZK-1 and (d) GluA2–STZGlu+CTZ. Note the widening of the ion channel pore and the unwinding of the pre-M4 helix in the open state structure.
TMD: The Gating Effector Domain
Conformational changes that originate in the LBDs are communicated by the LBD–TMD linkers to the TMD, where the ion channel pore opens or closes depending on the state of the LBD layer. In the closed state, the bundle crossing of the M3 helices occludes the ion permeation pathway, creating an upper gate, contributed by A617, A621, and T625 from all four subunits11 (Figure 6). Unique to subunits A and C is an extended M3 helix, where the side chains of M629 protrude toward the center of the pore and also contribute to the upper gate. This region of the pore is 2-fold symmetric, and in subunits B and D, M629 is part of the M3–S2 linker and does not contribute to the upper gate. Newly identified in the GluA2–GSG1LZK cryo-EM structure25 is a second, lower channel gate created by the Q/R site glutamines, Q586, which extend their side chains from the tips of the M2 loops toward the pore center. In neurons, brain compartment-specific mRNA editing of the GluA2 subunit Q/R site (glutamine to arginine) dramatically changes AMPAR function: the presence of arginine makes AMPARs Ca2+-impermeable and resistant to polyamine block.67−74 On the basis of the GluA2–GSG1LZK structure, replacement of the Q586 glutamine with arginine will place positively charged guanidinium groups in the middle of the ion channel pore, likely causing interference with Ca2+ permeation and polyamine block due to electrostatic repulsion.
For the channel to open, forces must be applied to the TMD via the LBD–TMD linkers. As discussed above, the changes in the LBD–TMD linkers are largely 2-fold symmetric, and therefore, it is not surprising that the changes at the top of the ion channel are also 2-fold symmetric25 (Movie 6 and Figure 6). Upon opening, the M3 helices in subunits A and C become one helical turn shorter, and methionines M629, which previously occluded the ion channel, flip away from the central pore axis. The major structural changes, however, occur in the M3 segments of subunits B and D, where a kink at A618 pulls the helices away from the pore axis, opening the upper gate for ion permeation. This A618 gating hinge, which is unique to tetrameric ion channels, is in the middle of “SYTANLAAF”, the most highly conserved motif in iGluRs.9 Therefore, we predict that the mechanism of ion channel opening will be largely similar across the entire iGluR family. At the lower gate, Q586 side chains flip outward from the ion permeation pathway upon channel opening and no longer occlude the pore. The cytoplasmic part of the selectivity filter, formed by extended regions of M2 and contributed to by the backbone carbonyls of Q587, G588, and C589, is better resolved in the open state GluA2–STZGlu+CTZ cryo-EM structure25 and appears to be more stable than in the closed channel structures. The pore seems to be wider here, as well, which is supported by the appearance of a clear density in the center of the pore, likely representing permeant sodium ions.25 Together, the conformational changes in the TMD suggest an iris-like mechanism of channel opening, similar to that of K+ channels, where the M3 pore-forming helices splay away from the central pore axis to allow for ion permeation (Movie 6).
The phenomenon of multiple conductance levels in AMPAR single-channel currents reflects changes in the pore dilation that are directly related to LBD occupancy by agonist and conformational changes in individual receptor subunits. We propose that the highest conductance level corresponds to the fully dilated pore, when the ion permeation is relieved at both channel gates by all four subunits. In turn, incomplete occupancy of LBDs by agonist results in asymmetric conformational changes in individual subunits, partial opening of the upper, lower, or both gates, and, as a result, incomplete dilation of the pore that can support ion permeation only at lower conductance levels.
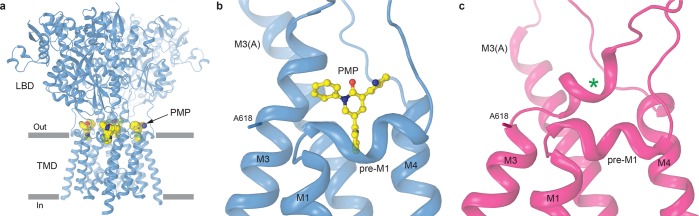
Interestingly, the structural changes that occur during ion channel opening in the AMPAR TMD and LBD–TMD linkers interfere with the binding of small-molecule noncompetitive inhibitors. These inhibitors, including perampanel (PMP, the only FDA-approved drug targeting AMPARs), GYKI-53655, and CP-465022, have been recently identified crystallographically to bind in the upper region of the TMD, termed the ion channel extracellular collar.17 Access to this site, formed by the pre-M1 helix and the extracellular portions of M1, M3, and M4, is open for noncompetitive inhibitor binding in the resting, closed state of the receptor (Figure 7a,b). In contrast, the upper portions of the kinked M3 helices in subunits B and D occlude access to the noncompetitive inhibitor binding sites in the open state (Figure 7c). This explains why the noncompetitive inhibitors preferentially interact with AMPARs in the closed state17,75 and supports the previously proposed inhibition mechanism,17 where the inhibitors occupying their binding sites act as “wedges” to prevent the conformational changes (kinking of the M3 helices at the A618 gating hinge in subunits B and D) associated with channel opening.25 Additionally, the rearrangements of the TMD collar region during gating explain why mutations around pre-M1 in both AMPARs and NMDARs have profound effects on gating and cause human neurological diseases.64,66,76−78
Figure 7.
Noncompetitive inhibitor binding site and gating. (a) Crystal structure of GluA2 in the closed, resting state bound to the noncompetitive inhibitor perampanel (PMP) (GluA2PMP, PDB entry 5L1F), with the ATD layer removed, viewed parallel to the membrane. The four PMP molecules bound in the ion channel extracellular collar are shown as space-filling models. (b) Close-up of the PMP binding site within GluA2PMP subunit B. The PMP molecule is shown in ball-and-stick representation. (c) Close-up of the same region in the open state GluA2–STZGlu+CTZ structure. The green asterisk indicates the region where PMP binds in GluA2PMP, which in the open state is occupied by the extracellular portion of M3 kinked at the A618 gating hinge.
ATD: The Gating Modulatory Domain
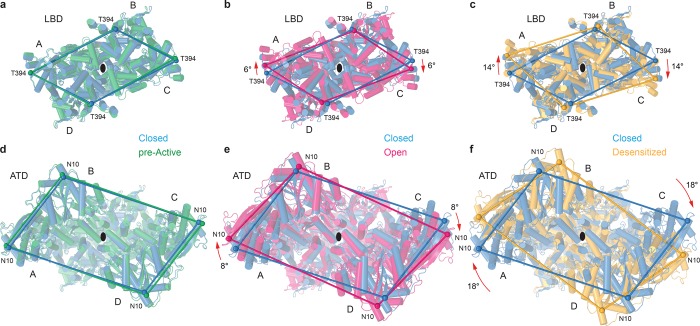
While the role of the ATD in AMPAR gating has remained mysterious, recent cryo-EM structures show that it certainly does not stand by idly; it appears to act as a modulator and communicator of the conformational changes that are happening in the receptor below. The ATD is connected to the LBD through the ATD–S1 linker, which is truncated in many structural studies to help reduce structural heterogeneity and flexibility. However, regardless of the wild-type or truncated state of this linker, early cryo-EM and X-ray crystallography studies put forth the idea that the ATDs would splay apart (i.e., rupture the cross-dimer interface) upon desensitization to help uncouple the ligand-bound state of the receptor from the channel. However, recent biophysical studies using cross-linking79 and fluorescence resonance energy transfer80 suggested that the ATDs maintain their tetrameric assembly and do not splay apart during gating. Indeed, the recent crystal16 and cryo-EM25,26 structures show that the changes in the LBDs described above alter the top of the LBD layer (Movie 7 and Figure 8), resulting in rigid-body movement of the entire ATD tetramer during gating (Movie 8 and Figure 8). During the C to PA transition, there is little overall movement of the LBD tetramer (Figure 8a), and thus, the ATD layer stays nearly the same (Figure 8d). However, upon activation and channel opening, the tops of the A and C subunit LBDs rotate by 6° (Figure 8b), resulting in a rigid-body rotation of the entire ATD tetramer by 8° (Figure 8e). Even larger changes are observed during desensitization, when a 14° rotation of the tops of the A and C subunit LBDs (Figure 8c) results in a dramatic 18° rigid-body rotation of the entire ATD tetramer (Figure 8f). Consequently, profound changes in the LBD layer domain arrangement and interfaces during gating do not seem to alter the tetrameric arrangement of AMPAR ATDs. The entire ATD layer rotates as a rigid body (Figure 8d–f), not only emphasizing the important role of the ATD in AMPAR assembly7 but also providing a way for modulating AMPAR function through interaction with trans-synaptic molecular elements.81,82
Figure 8.
Overall movements of the LBD and ATD tetramers during gating. Superposition of (a–c) LBD and (d–f) ATD tetramers from GluA2–GSG1LZK-1 (blue, closed state) and (a and d) GluA2NOW (green, pre-active state), (b and e) GluA2–STZGlu+CTZ (magenta, open state), or (c and f) GluA2–GSG1LQuis (orange, desensitized state) viewed extracellularly along the axis of the overall 2-fold receptor symmetry (black ovals). Cα atoms of T394 (a–c) and N10 (d–f) are shown as spheres of the corresponding color, connected by straight lines. Relative motions of the top portions of the LBDs and rigid-body rotations of the ATD tetramer are indicated by red arrows.
We hypothesize that the ATD splaying observed in earlier crystallographic and cryo-EM studies13,18,83 is an artifact of the sample preparation and treatment, as receptors with wild-type linkers also maintain their ATD layer integrity during desensitization, as evidenced by cryo-EM data from a recent study.49 In addition, the splaying apart of ATDs unlikely happens in vivo, given the crowded synaptic space84 and numerous cross-synaptic interactions that modulate iGluR function.85−87
Conclusions and Outlook
A couple of years ago the structural determinants of iGluR gating remained ambiguous. Now, a collection of recent crystallography and cryo-EM AMPAR structures allow structural visualization of the entire iGluR gating mechanism for the first time (Movies 1–9). However, many key questions remain unanswered. How conserved is the AMPAR gating mechanism in different iGluR subtypes? What are the structural determinants of multiple conductance levels? What is the exact structural mechanism of iGluR assembly? What are structural mechanisms of small-molecule interactions with the gating machinery of iGluRs? What are the energetic determinants along the iGluR gating transitions? How do different auxiliary subunits affect the structural mechanisms of iGluR gating? What are the structures of iGluR CTDs, and how do these domains contribute to receptor function in neurons? We expect the coming years will be an exciting time to address these and many other questions about iGluR structure and function and that building upon the recent structures not only enhances our understanding of the molecular bases of excitatory neurotransmission but also will contribute to drug design targeting iGluRs in neurological diseases.
Acknowledgments
We thank M. V. Yelshanskaya and L. L. McGoldrick for valuable comments and edits on this Perspective.
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.biochem.7b00891.
E.C.T. is supported by National Institutes of Health (NIH) Grant F31 NS093838. A.I.S. is supported by NIH Grants R01 NS083660 and R01 CA206573, the Amgen Young Investigator Award, and the Irma T. Hirschl Career Scientist Award.
The authors declare no competing financial interest.
Supplementary Material
References
- Traynelis S. F.; Wollmuth L. P.; McBain C. J.; Menniti F. S.; Vance K. M.; Ogden K. K.; Hansen K. B.; Yuan H.; Myers S. J.; Dingledine R.; Sibley D. (2010) Glutamate receptor ion channels: structure, regulation, and function. Pharmacol. Rev. 62, 405–496. 10.1124/pr.109.002451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K.; Goodman L.; Fourie C.; Schenk S.; Leitch B.; Montgomery J. M. (2016) AMPA Receptors as Therapeutic Targets for Neurological Disorders. Adv. Protein Chem. Struct. Biol. 103, 203–261. 10.1016/bs.apcsb.2015.10.004. [DOI] [PubMed] [Google Scholar]
- Zhou Q.; Sheng M. (2013) NMDA receptors in nervous system diseases. Neuropharmacology 74, 69–75. 10.1016/j.neuropharm.2013.03.030. [DOI] [PubMed] [Google Scholar]
- Lerma J.; Marques J. M. (2013) Kainate receptors in health and disease. Neuron 80, 292–311. 10.1016/j.neuron.2013.09.045. [DOI] [PubMed] [Google Scholar]
- Wo Z. G.; Oswald R. E. (1995) Unraveling the modular design of glutamate-gated ion channels. Trends Neurosci. 18, 161–168. 10.1016/0166-2236(95)93895-5. [DOI] [PubMed] [Google Scholar]
- Sobolevsky A. I. (2015) Structure and gating of tetrameric glutamate receptors. J. Physiol. 593, 29–38. 10.1113/jphysiol.2013.264911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayalon G.; Stern-Bach Y. (2001) Functional assembly of AMPA and kainate receptors is mediated by several discrete protein-protein interactions. Neuron 31, 103–113. 10.1016/S0896-6273(01)00333-6. [DOI] [PubMed] [Google Scholar]
- Stern-Bach Y.; Bettler B.; Hartley M.; Sheppard P. O.; O’Hara P. J.; Heinemann S. F. (1994) Agonist selectivity of glutamate receptors is specified by two domains structurally related to bacterial amino acid-binding proteins. Neuron 13, 1345–1357. 10.1016/0896-6273(94)90420-0. [DOI] [PubMed] [Google Scholar]
- Wollmuth L. P.; Sobolevsky A. I. (2004) Structure and gating of the glutamate receptor ion channel. Trends Neurosci. 27, 321–328. 10.1016/j.tins.2004.04.005. [DOI] [PubMed] [Google Scholar]
- Sheng M.; Pak D. T. (2000) Ligand-gated ion channel interactions with cytoskeletal and signaling proteins. Annu. Rev. Physiol. 62, 755–778. 10.1146/annurev.physiol.62.1.755. [DOI] [PubMed] [Google Scholar]
- Sobolevsky A. I.; Rosconi M. P.; Gouaux E. (2009) X-ray structure, symmetry and mechanism of an AMPA-subtype glutamate receptor. Nature 462, 745–756. 10.1038/nature08624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L.; Durr K. L.; Gouaux E. (2014) X-ray structures of AMPA receptor-cone snail toxin complexes illuminate activation mechanism. Science 345, 1021–1026. 10.1126/science.1258409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durr K. L.; Chen L.; Stein R. A.; De Zorzi R.; Folea I. M.; Walz T.; McHaourab H. S.; Gouaux E. (2014) Structure and Dynamics of AMPA Receptor GluA2 in Resting, Pre-Open, and Desensitized States. Cell 158, 778–792. 10.1016/j.cell.2014.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karakas E.; Furukawa H. (2014) Crystal structure of a heterotetrameric NMDA receptor ion channel. Science 344, 992–997. 10.1126/science.1251915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C. H.; Lu W.; Michel J. C.; Goehring A.; Du J.; Song X.; Gouaux E. (2014) NMDA receptor structures reveal subunit arrangement and pore architecture. Nature 511, 191–197. 10.1038/nature13548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yelshanskaya M. V.; Li M.; Sobolevsky A. I. (2014) Structure of an agonist-bound ionotropic glutamate receptor. Science 345, 1070–1074. 10.1126/science.1256508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yelshanskaya M. V.; Singh A. K.; Sampson J. M.; Narangoda C.; Kurnikova M.; Sobolevsky A. I. (2016) Structural Bases of Noncompetitive Inhibition of AMPA-Subtype Ionotropic Glutamate Receptors by Antiepileptic Drugs. Neuron 91, 1305–1315. 10.1016/j.neuron.2016.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyerson J. R.; Kumar J.; Chittori S.; Rao P.; Pierson J.; Bartesaghi A.; Mayer M. L.; Subramaniam S. (2014) Structural mechanism of glutamate receptor activation and desensitization. Nature 514, 328–334. 10.1038/nature13603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyerson J. R.; Chittori S.; Merk A.; Rao P.; Han T. H.; Serpe M.; Mayer M. L.; Subramaniam S. (2016) Structural basis of kainate subtype glutamate receptor desensitization. Nature 537, 567–571. 10.1038/nature19352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Twomey E. C.; Yelshanskaya M. V.; Grassucci R. A.; Frank J.; Sobolevsky A. I. (2016) Elucidation of AMPA receptor-stargazin complexes by cryo-electron microscopy. Science 353, 83–86. 10.1126/science.aaf8411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y.; Chen S.; Yoshioka C.; Baconguis I.; Gouaux E. (2016) Architecture of fully occupied GluA2 AMPA receptor-TARP complex elucidated by cryo-EM. Nature 536, 108–111. 10.1038/nature18961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu S.; Stein R. A.; Yoshioka C.; Lee C. H.; Goehring A.; McHaourab H. S.; Gouaux E. (2016) Mechanism of NMDA Receptor Inhibition and Activation. Cell 165, 704–714. 10.1016/j.cell.2016.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajima N.; Karakas E.; Grant T.; Simorowski N.; Diaz-Avalos R.; Grigorieff N.; Furukawa H. (2016) Activation of NMDA receptors and the mechanism of inhibition by ifenprodil. Nature 534, 63–68. 10.1038/nature17679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu W.; Du J.; Goehring A.; Gouaux E. (2017) Cryo-EM structures of the triheteromeric NMDA receptor and its allosteric modulation. Science 355, eaal3729. 10.1126/science.aal3729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Twomey E. C.; Yelshanskaya M. V.; Grassucci R. A.; Frank J.; Sobolevsky A. I. (2017) Channel opening and gating mechanism in AMPA-subtype glutamate receptors. Nature 549, 60–65. 10.1038/nature23479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Twomey E. C.; Yelshanskaya M. V.; Grassucci R. A.; Frank J.; Sobolevsky A. I. (2017) Structural Bases of Desensitization in AMPA Receptor-Auxiliary Subunit Complexes. Neuron 94, 569–580. 10.1016/j.neuron.2017.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert A.; Howe J. R. (2003) How AMPA receptor desensitization depends on receptor occupancy. J. Neurosci. 23, 847–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popescu G.; Auerbach A. (2003) Modal gating of NMDA receptors and the shape of their synaptic response. Nat. Neurosci. 6, 476–483. 10.1038/nn1044. [DOI] [PubMed] [Google Scholar]
- Vance K. M.; Hansen K. B.; Traynelis S. F. (2013) Modal gating of GluN1/GluN2D NMDA receptors. Neuropharmacology 71, 184–190. 10.1016/j.neuropharm.2013.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poon K.; Ahmed A. H.; Nowak L. M.; Oswald R. E. (2011) Mechanisms of modal activation of GluA3 receptors. Mol. Pharmacol. 80, 49–59. 10.1124/mol.111.071688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cull-Candy S. G.; Usowicz M. M. (1987) Multiple-conductance channels activated by excitatory amino acids in cerebellar neurons. Nature 325, 525–528. 10.1038/325525a0. [DOI] [PubMed] [Google Scholar]
- Jahr C. E.; Stevens C. F. (1987) Glutamate activates multiple single channel conductances in hippocampal neurons. Nature 325, 522–525. 10.1038/325522a0. [DOI] [PubMed] [Google Scholar]
- Rosenmund C.; Stern-Bach Y.; Stevens C. F. (1998) The tetrameric structure of a glutamate receptor channel,. Science 280, 1596–1599. 10.1126/science.280.5369.1596. [DOI] [PubMed] [Google Scholar]
- Smith T. C.; Howe J. R. (2000) Concentration-dependent substate behavior of native AMPA receptors. Nat. Neurosci. 3, 992–997. 10.1038/79931. [DOI] [PubMed] [Google Scholar]
- Prieto M. L.; Wollmuth L. P. (2010) Gating modes in AMPA receptors. J. Neurosci. 30, 4449–4459. 10.1523/JNEUROSCI.5613-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin R.; Banke T. G.; Mayer M. L.; Traynelis S. F.; Gouaux E. (2003) Structural basis for partial agonist action at ionotropic glutamate receptors. Nat. Neurosci. 6, 803–810. 10.1038/nn1091. [DOI] [PubMed] [Google Scholar]
- Tomita S.; Adesnik H.; Sekiguchi M.; Zhang W.; Wada K.; Howe J. R.; Nicoll R. A.; Bredt D. S. (2005) Stargazin modulates AMPA receptor gating and trafficking by distinct domains. Nature 435, 1052–1058. 10.1038/nature03624. [DOI] [PubMed] [Google Scholar]
- Jackson A. C.; Nicoll R. A. (2011) The expanding social network of ionotropic glutamate receptors: TARPs and other transmembrane auxiliary subunits,. Neuron 70, 178–199. 10.1016/j.neuron.2011.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howe J. R. (2015) Modulation of non-NMDA receptor gating by auxiliary subunits. J. Physiol. 593, 61–72. 10.1113/jphysiol.2014.273904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haering S. C.; Tapken D.; Pahl S.; Hollmann M. (2014) Auxiliary subunits: shepherding AMPA receptors to the plasma membrane. Membranes (Basel, Switz.) 4, 469–490. 10.3390/membranes4030469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bettler B.; Fakler B. (2017) Ionotropic AMPA-type glutamate and metabotropic GABAB receptors: determining cellular physiology by proteomes. Curr. Opin. Neurobiol. 45, 16–23. 10.1016/j.conb.2017.02.011. [DOI] [PubMed] [Google Scholar]
- Shanks N. F.; Savas J. N.; Maruo T.; Cais O.; Hirao A.; Oe S.; Ghosh A.; Noda Y.; Greger I. H.; Yates J. R. 3rd; Nakagawa T. (2012) Differences in AMPA and kainate receptor interactomes facilitate identification of AMPA receptor auxiliary subunit GSG1L. Cell Rep. 1, 590–598. 10.1016/j.celrep.2012.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwenk J.; Harmel N.; Brechet A.; Zolles G.; Berkefeld H.; Muller C. S.; Bildl W.; Baehrens D.; Huber B.; Kulik A.; Klocker N.; Schulte U.; Fakler B. (2012) High-resolution proteomics unravel architecture and molecular diversity of native AMPA receptor complexes. Neuron 74, 621–633. 10.1016/j.neuron.2012.03.034. [DOI] [PubMed] [Google Scholar]
- McGee T. P.; Bats C.; Farrant M.; Cull-Candy S. G. (2015) Auxiliary Subunit GSG1L Acts to Suppress Calcium-Permeable AMPA Receptor Function. J. Neurosci. 35, 16171–16179. 10.1523/JNEUROSCI.2152-15.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu X.; Mao X.; Lussier M. P.; Hutchison M. A.; Zhou L.; Hamra F. K.; Roche K. W.; Lu W. (2016) GSG1L suppresses AMPA receptor-mediated synaptic transmission and uniquely modulates AMPA receptor kinetics in hippocampal neurons. Nat. Commun. 7, 10873. 10.1038/ncomms10873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W.; Robert A.; Vogensen S. B.; Howe J. R. (2006) The relationship between agonist potency and AMPA receptor kinetics,. Biophys. J. 91, 1336–1346. 10.1529/biophysj.106.084426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Priel A.; Kolleker A.; Ayalon G.; Gillor M.; Osten P.; Stern-Bach Y. (2005) Stargazin reduces desensitization and slows deactivation of the AMPA-type glutamate receptors. J. Neurosci. 25, 2682–2686. 10.1523/JNEUROSCI.4834-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yelshanskaya M. V.; Singh A. K.; Sampson J. M.; Narangoda C.; Kurnikova M.; Sobolevsky A. I. (2016) Structural bases of noncompetitive inhibition of AMPA subtype ionotropic glutamate receptors by antiepileptic drugs. Neuron 91, 1305–1315. 10.1016/j.neuron.2016.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S.; Zhao Y.; Wang Y.; Shekhar M.; Tajkhorshid E.; Gouaux E. (2017) Activation and Desensitization Mechanism of AMPA Receptor-TARP Complex by Cryo-EM. Cell 170, 1234–1246. 10.1016/j.cell.2017.07.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armstrong N.; Gouaux E. (2000) Mechanisms for activation and antagonism of an AMPA-sensitive glutamate receptor: crystal structures of the GluR2 ligand binding core. Neuron 28, 165–181. 10.1016/S0896-6273(00)00094-5. [DOI] [PubMed] [Google Scholar]
- Maltsev A. S.; Ahmed A. H.; Fenwick M. K.; Jane D. E.; Oswald R. E. (2008) Mechanism of partial agonism at the GluR2 AMPA receptor: Measurements of lobe orientation in solution. Biochemistry 47, 10600–10610. 10.1021/bi800843c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W.; Cho Y.; Lolis E.; Howe J. R. (2008) Structural and single-channel results indicate that the rates of ligand binding domain closing and opening directly impact AMPA receptor gating. J. Neurosci. 28, 932–943. 10.1523/JNEUROSCI.3309-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed A. H.; Wang S.; Chuang H. H.; Oswald R. E. (2011) Mechanism of AMPA receptor activation by partial agonists: disulfide trapping of closed lobe conformations. J. Biol. Chem. 286, 35257–35266. 10.1074/jbc.M111.269001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maclean D. M.; Wong A. Y.; Fay A. M.; Bowie D. (2011) Cations but not anions regulate the responsiveness of kainate receptors. J. Neurosci. 31, 2136–2144. 10.1523/JNEUROSCI.4314-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau A. Y.; Roux B. (2011) The hidden energetics of ligand binding and activation in a glutamate receptor. Nat. Struct. Mol. Biol. 18, 283–287. 10.1038/nsmb.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landes C. F.; Rambhadran A.; Taylor J. N.; Salatan F.; Jayaraman V. (2011) Structural landscape of isolated agonist-binding domains from single AMPA receptors. Nat. Chem. Biol. 7, 168–173. 10.1038/nchembio.523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armstrong N.; Jasti J.; Beich-Frandsen M.; Gouaux E. (2006) Measurement of conformational changes accompanying desensitization in an ionotropic glutamate receptor. Cell 127, 85–97. 10.1016/j.cell.2006.08.037. [DOI] [PubMed] [Google Scholar]
- Sun Y.; Olson R.; Horning M.; Armstrong N.; Mayer M.; Gouaux E. (2002) Mechanism of glutamate receptor desensitization. Nature 417, 245–253. 10.1038/417245a. [DOI] [PubMed] [Google Scholar]
- Dong H.; Zhou H. X. (2011) Atomistic mechanism for the activation and desensitization of an AMPA-subtype glutamate receptor. Nat. Commun. 2, 354. 10.1038/ncomms1362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazi R.; Dai J.; Sweeney C.; Zhou H. X.; Wollmuth L. P. (2014) Mechanical coupling maintains the fidelity of NMDA receptor-mediated currents. Nat. Neurosci. 17, 914–922. 10.1038/nn.3724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gan Q.; Dai J.; Zhou H. X.; Wollmuth L. P. (2016) The Transmembrane Domain Mediates Tetramerization of alpha-Amino-3-hydroxy-5-methyl-4-isoxazolepropionic Acid (AMPA) Receptors,. J. Biol. Chem. 291, 6595–6606. 10.1074/jbc.M115.686246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salussolia C. L.; Corrales A.; Talukder I.; Kazi R.; Akgul G.; Bowen M.; Wollmuth L. P. (2011) Interaction of the M4 segment with other transmembrane segments is required for surface expression of mammalian alpha-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid (AMPA) receptors. J. Biol. Chem. 286, 40205–40218. 10.1074/jbc.M111.268839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salussolia C. L.; Gan Q.; Kazi R.; Singh P.; Allopenna J.; Furukawa H.; Wollmuth L. P. (2013) A eukaryotic specific transmembrane segment is required for tetramerization in AMPA receptors,. J. Neurosci. 33, 9840–9845. 10.1523/JNEUROSCI.2626-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan H.; Low C. M.; Moody O. A.; Jenkins A.; Traynelis S. F. (2015) Ionotropic GABA and Glutamate Receptor Mutations and Human Neurologic Diseases. Mol. Pharmacol. 88, 203–217. 10.1124/mol.115.097998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amin J. B.; Salussolia C. L.; Chan K.; Regan M. C.; Dai J.; Zhou H. X.; Furukawa H.; Bowen M. E.; Wollmuth L. P. (2017) Divergent roles of a peripheral transmembrane segment in AMPA and NMDA receptors. J. Gen. Physiol. 149, 661–680. 10.1085/jgp.201711762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yelshanskaya M. V.; Mesbahi-Vasey S.; Kurnikova M. G.; Sobolevsky A. I. (2017) Role of the Ion Channel Extracellular Collar in AMPA Receptor Gating. Sci. Rep. 7, 1050. 10.1038/s41598-017-01146-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommer B.; Kohler M.; Sprengel R.; Seeburg P. H. (1991) RNA editing in brain controls a determinant of ion flow in glutamate-gated channels. Cell 67, 11–19. 10.1016/0092-8674(91)90568-J. [DOI] [PubMed] [Google Scholar]
- Burnashev N.; Schoepfer R.; Monyer H.; Ruppersberg J. P.; Gunther W.; Seeburg P. H.; Sakmann B. (1992) Control by asparagine residues of calcium permeability and magnesium blockade in the NMDA receptor. Science 257, 1415–1419. 10.1126/science.1382314. [DOI] [PubMed] [Google Scholar]
- Bowie D.; Mayer M. L. (1995) Inward rectification of both AMPA and kainate subtype glutamate receptors generated by polyamine-mediated ion channel block. Neuron 15, 453–462. 10.1016/0896-6273(95)90049-7. [DOI] [PubMed] [Google Scholar]
- Donevan S. D.; Rogawski M. A. (1995) Intracellular polyamines mediate inward rectification of Ca(2+)-permeable alpha-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid receptors. Proc. Natl. Acad. Sci. U. S. A. 92, 9298–9302. 10.1073/pnas.92.20.9298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isa T.; Iino M.; Itazawa S.; Ozawa S. (1995) Spermine mediates inward rectification of Ca(2+)-permeable AMPA receptor channels. NeuroReport 6, 2045–2048. 10.1097/00001756-199510010-00022. [DOI] [PubMed] [Google Scholar]
- Kamboj S. K.; Swanson G. T.; Cull-Candy S. G. (1995) Intracellular spermine confers rectification on rat calcium-permeable AMPA and kainate receptors. J. Physiol. 486, 297–303. 10.1113/jphysiol.1995.sp020812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koh D. S.; Burnashev N.; Jonas P. (1995) Block of native Ca(2+)-permeable AMPA receptors in rat brain by intracellular polyamines generates double rectification. J. Physiol. 486, 305–312. 10.1113/jphysiol.1995.sp020813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huettner J. E. (2015) Glutamate receptor pores. J. Physiol. 593, 49–59. 10.1113/jphysiol.2014.272724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balannik V.; Menniti F. S.; Paternain A. V.; Lerma J.; Stern-Bach Y. (2005) Molecular mechanism of AMPA receptor noncompetitive antagonism. Neuron 48, 279–288. 10.1016/j.neuron.2005.09.024. [DOI] [PubMed] [Google Scholar]
- Ogden K. K.; Traynelis S. F. (2013) Contribution of the M1 transmembrane helix and pre-M1 region to positive allosteric modulation and gating of N-methyl-D-aspartate receptors. Mol. Pharmacol. 83, 1045–1056. 10.1124/mol.113.085209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogden K. K.; Chen W.; Swanger S. A.; McDaniel M. J.; Fan L. Z.; Hu C.; Tankovic A.; Kusumoto H.; Kosobucki G. J.; Schulien A. J.; Su Z.; Pecha J.; Bhattacharya S.; Petrovski S.; Cohen A. E.; Aizenman E.; Traynelis S. F.; Yuan H. (2017) Molecular Mechanism of Disease-Associated Mutations in the Pre-M1 Helix of NMDA Receptors and Potential Rescue Pharmacology. PLoS Genet. 13, e1006536 10.1371/journal.pgen.1006536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alsaloum M.; Kazi R.; Gan Q.; Amin J.; Wollmuth L. P. (2016) A Molecular Determinant of Subtype-Specific Desensitization in Ionotropic Glutamate Receptors,. J. Neurosci. 36, 2617–2622. 10.1523/JNEUROSCI.2667-15.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yelshanskaya M. V.; Saotome K.; Singh A. K.; Sobolevsky A. I. (2016) Probing Intersubunit Interfaces in AMPA-subtype Ionotropic Glutamate Receptors. Sci. Rep. 6, 19082. 10.1038/srep19082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaikh S. A.; Dolino D. M.; Lee G.; Chatterjee S.; MacLean D. M.; Flatebo C.; Landes C. F.; Jayaraman V. (2016) Stargazin Modulation of AMPA Receptors. Cell Rep. 17, 328–335. 10.1016/j.celrep.2016.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson J. F.; Ho H.; Greger I. H. (2017) Synaptic transmission and plasticity require AMPA receptor anchoring via its N-terminal domain. eLife 6, 23024. 10.7554/eLife.23024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Nafria J.; Herguedas B.; Watson J. F.; Greger I. H. (2016) The dynamic AMPA receptor extracellular region: a platform for synaptic protein interactions,. J. Physiol. 594, 5449–5458. 10.1113/JP271844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa T.; Cheng Y.; Ramm E.; Sheng M.; Walz T. (2005) Structure and different conformational states of native AMPA receptor complexes. Nature 433, 545–549. 10.1038/nature03328. [DOI] [PubMed] [Google Scholar]
- Lee S. H.; Jin C.; Cai E.; Ge P.; Ishitsuka Y.; Teng K. W.; de Thomaz A. A.; Nall D.; Baday M.; Jeyifous O.; Demonte D.; Dundas C. M.; Park S.; Delgado J. Y.; Green W. N.; Selvin P. R. (2017) Super-resolution imaging of synaptic and Extra-synaptic AMPA receptors with different-sized fluorescent probes. eLife 6, 27744. 10.7554/eLife.27744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverman J. B.; Restituito S.; Lu W.; Lee-Edwards L.; Khatri L.; Ziff E. B. (2007) Synaptic anchorage of AMPA receptors by cadherins through neural plakophilin-related arm protein AMPA receptor-binding protein complexes. J. Neurosci. 27, 8505–8516. 10.1523/JNEUROSCI.1395-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elegheert J.; Kakegawa W.; Clay J. E.; Shanks N. F.; Behiels E.; Matsuda K.; Kohda K.; Miura E.; Rossmann M.; Mitakidis N.; Motohashi J.; Chang V. T.; Siebold C.; Greger I. H.; Nakagawa T.; Yuzaki M.; Aricescu A. R. (2016) Structural basis for integration of GluD receptors within synaptic organizer complexes. Science 353, 295–299. 10.1126/science.aae0104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda K.; Budisantoso T.; Mitakidis N.; Sugaya Y.; Miura E.; Kakegawa W.; Yamasaki M.; Konno K.; Uchigashima M.; Abe M.; Watanabe I.; Kano M.; Watanabe M.; Sakimura K.; Aricescu A. R.; Yuzaki M. (2016) Transsynaptic Modulation of Kainate Receptor Functions by C1q-like Proteins. Neuron 90, 752–767. 10.1016/j.neuron.2016.04.001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.