Figure 1.
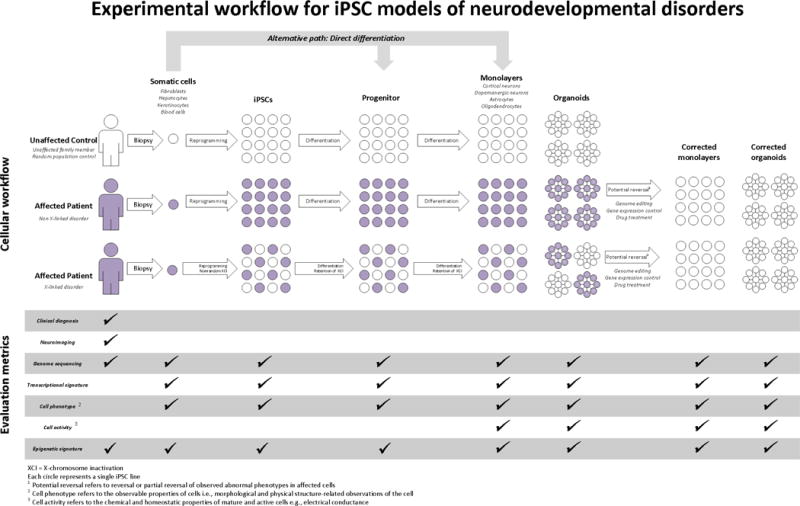
Experimental workflow for hiPSC models of neurodevelopmental disorders. Different experimental options are shown with regards to type of controls (cross-sectional, matched pair or family contrpol), choice of reprogrammed cell type, type of differentiation protocol, and outcome metrics. For patients with X-linked disorders, different colored cells represent cells with either the wild type or the mutated X allele. Corrected cells represent the same patient-derived cells after genome editing or drug treatment.
