Figure 1.
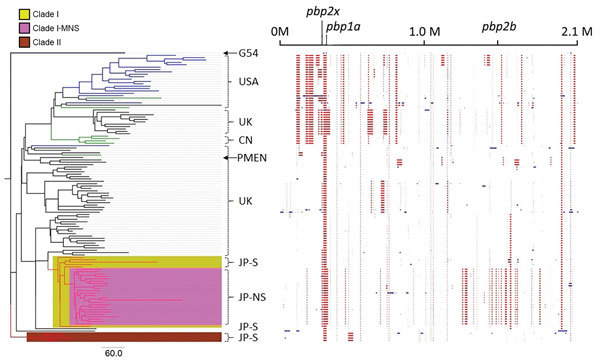
Phylogenic tree and predicted recombination sites created in Genealogies Unbiased By recomBinations In Nucleotide Sequences (28) by using all Japan and global serotype 15A-ST63 pneumococcal isolates. Branch colors in the tree indicate where the isolates were collected: red, Japan; black, United Kingdom; blue, United States; green, Canada. The column on the right of the tree indicates the main region from which the isolates were derived, meropenem susceptibility, and isolate names. The phylogenic tree was created by using Streptococcus pneumoniae G54 as an outgroup isolate. Clade I consists of only Japan serotype 15A-ST63 isolates; clade I-MNS consists of only Japan meropenem-nonsusceptible serotype 15A-ST63 isolates; clade II consists of the rest of the Japan meropenem-susceptible serotype 15A-ST63 isolates that are not included in clade I. The block chart on the right shows the predicted recombination sites in each isolate. Blue blocks are unique to a single isolate; red blocks are shared by multiple isolates. All isolates shaded in pink are meropenem nonsusceptible. Arrows indicate reference strains S. pneumoniae G54 and PMEN 15A-25. Scale bar indicates nucleotide substitutions per site; CN, Canada; G54, S. pneumoniae G54; M, million base pairs; JP-NS, Japan meropenem nonsusceptible; JP-S, Japan meropenem susceptible; PMEN, Pneumococcal Molecular Epidemiology Network; ST, sequence type; UK, United Kingdom; USA, United States.
