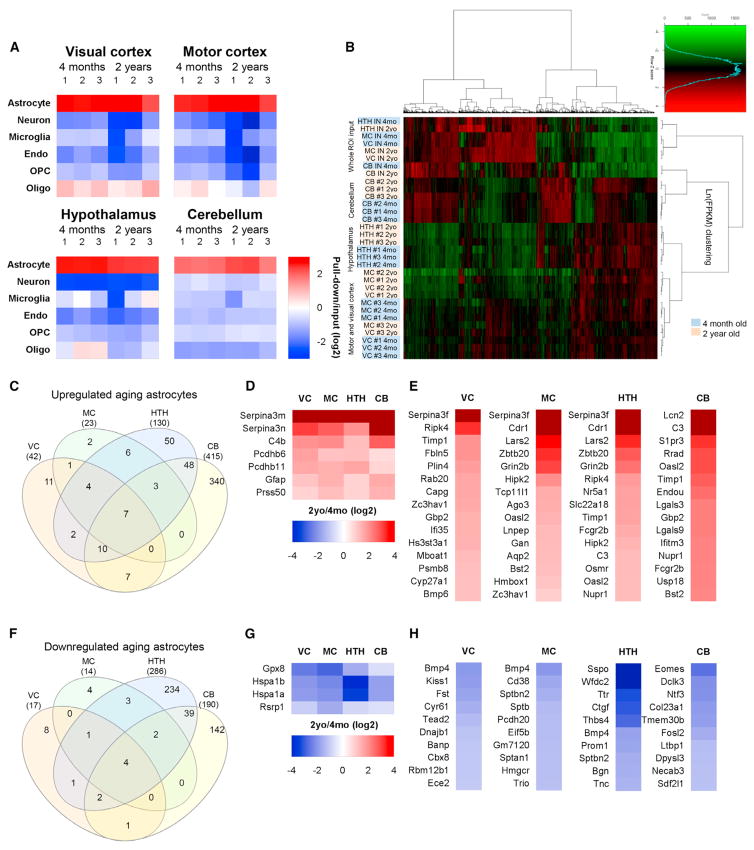
Figure 2. Purification and RNA-Seq of Astrocyte-Enriched mRNA Identifies Differentially Expressed Genes between Adult and Aging Astrocytes.
(A) Analysis of RNA-seq data for cell-specific mRNA demonstrates enrichment for astrocytes over other cell types in the pull-down (astrocyte mRNA) compared with input (total mRNA) (see Table S1 for the gene list).
(B) Clustering analysis of the top 4,401 expressed genes shows that astrocyte-ribotag pull-down samples cluster away from input mRNA; samples cluster by region of isolation and, within region, by age. n = 3 mice for each brain region and age (astrocyte samples). See Table S2 for FPKM for all genes.
(C) Venn diagram showing overlap of genes upregulated in astrocytes from 4 different brain regions between 4 months and 2 years.
(D) Heatmap of genes that are significantly upregulated in astrocytes by aging in all regions.
(E) Heatmaps of top 15 genes upregulated in astrocytes in each brain region by aging (excluding those in D; see Tables S3 and S4 for a full list).
(F) Venn diagram of genes downregulated in astrocytes from 4 different brain regions between 4 months and 2 years.
(G) Heatmap of genes that are significantly downregulated in astrocytes by aging in all regions.
(H) Heatmaps of the top 10 genes downregulated in astrocytes in each brain region by aging (excluding those in G; see Tables S3 and S4 for a full list). All heatmaps presented as fold change 2 years/4 months (log2). See also Figure S1 and Tables S1, S2, S3, and S4.