Figure 1. Determining the full length of the transcript found within the HBS1L-MYB intergenic region.
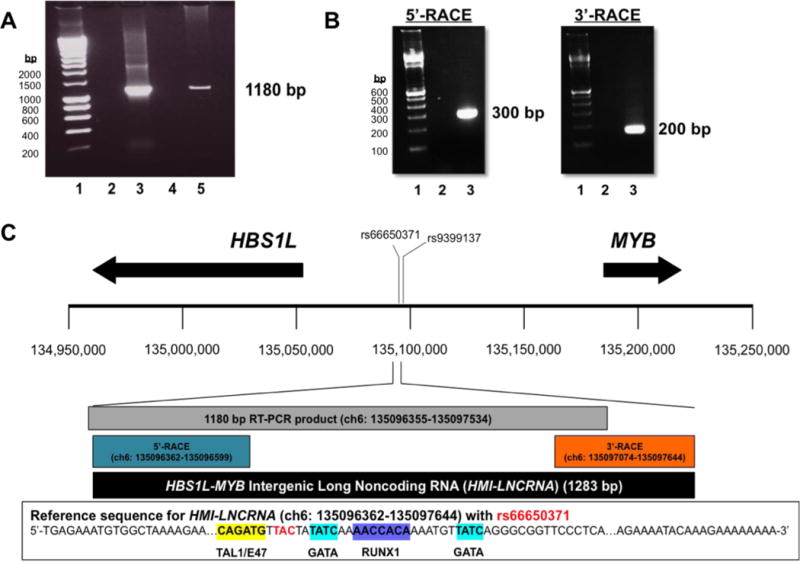
(A) PCR was done to amplify a 1180 bp region within the HBS1L-MYB intergenic region using cDNA from K652 cells. Samples were run on agarose gels. Lane 1: 100-bp DNA ladder; lane 2: negative control, PCR mix without DNA input; lane 3: positive control, PCR mix with genomic DNA; lane 4: PCR mix with cDNA generated without reverse transcriptase; and Lane 5: PCR mix with cDNA generated with reverse transcriptase. (B) Agarose gels show PCR products from the 5′-RACE reactions (using RNA from thymus) and 3′-RACE reactions (using RNA from K562 cells). For 5′-RACE, lane 1: 100-bp DNA ladder; lane 2: PCR mix with cDNA generated without Tobacco Acid Pyrophosphatase (TAP) treatment; and lane 3: PCR mix with cDNA generated with TAP treatment. For 3′-RACE, lane 1: 100-bp DNA ladder; lane 2: PCR mix with cDNA generated without reverse transcriptase; and lane 3: PCR mix with cDNA generated with reverse transcriptase. (C) Illustration of genomic region between chr6: 134,950,000–135,250,000 (hg38), showing approximate locations of the 3-bp deletion polymorphism rs66650371 and rs9399137 a SNP in LD with rs66650371, and HBS1L and MYB (arrows represent transcription direction and approximate length of genes). Based on DNA sequencing of RACE products, the 5′- and 3′-ends of the transcript were revealed to determine the full length of the transcript, which is 1283 bp in length and named the HBS1L-MYB Intergenic Long Noncoding RNA (HMI-LNCRNA). Located in the genomic sequence for HMI-LNCRNA are binding sites for erythroid-specific transcription factors TAL1/E47, GATA and RUNX1, and rs66650371. HMI-LNCRNA does not include rs9399137.
