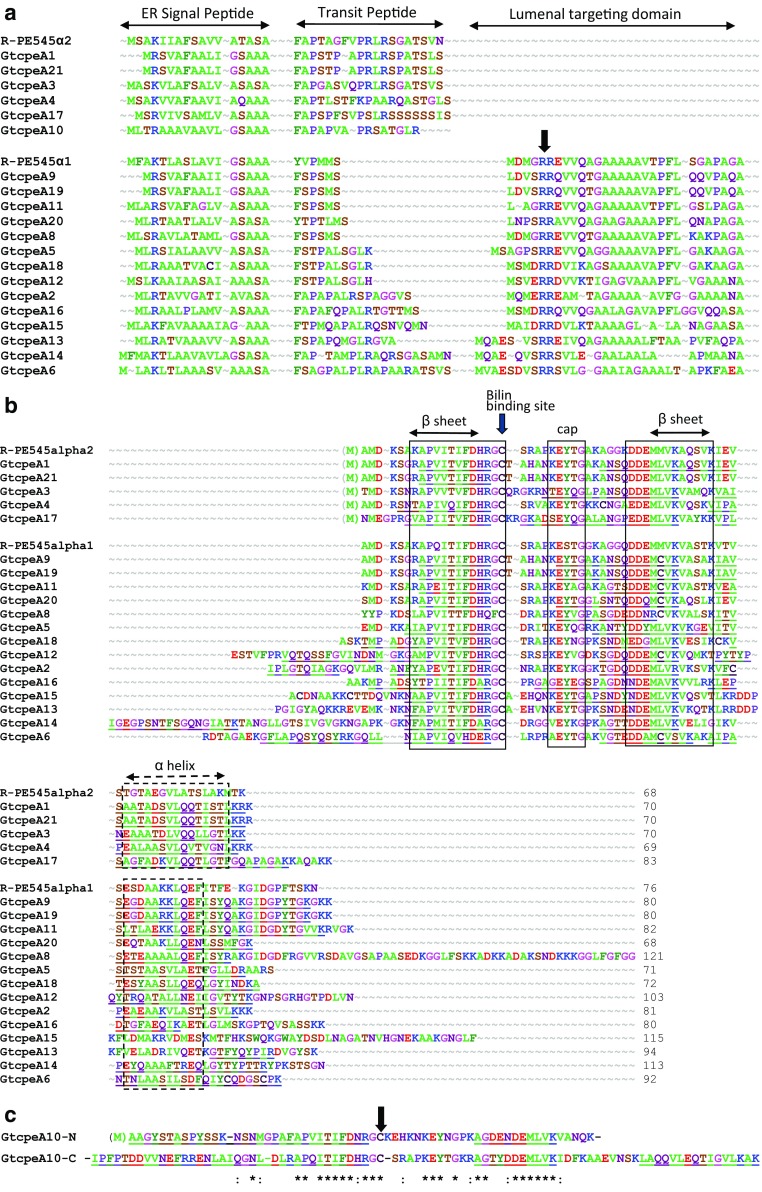
Fig. 4.
Multiple sequence alignment of the G. theta α-subunit sequences colored according to chemical similarity. a Precursor targeting sequences. All precursors start with an ER signal peptide, cleaved from the following chloroplast transit peptide at the conserved motif AXA_(F/Y) (Gruber et al. 2007). The C-terminal cleavage site of the chloroplast transit peptide is predicted according to Huesgen et al. (2013) with a conserved Leu or Met at the -2 position. Where present, the lumenal targeting domain has a twin arginine motif near the N-terminus (arrow) and a hydrophobic core, typical of proteins targeted to the thylakoid lumen via the TAT import machinery, and a C-terminal AXA cleavage site to release the mature protein. b Mature proteins aligned with the α1- and α2-sequences from the crystal structure of PE545 from Rhodomonas sp. CS24 (Wilk et al. 1999). Peptides identified by mass spectrometry are underlined. N-terminal Met are in parenthesis to indicate the high probability that they are removed by Met aminopeptidase (Huesgen et al. 2013; reviewed by Giglione and Meinnel 2001). Highly conserved regions corresponding to secondary structural elements of the Rhodomonas structure are boxed. The EYxG motif forms a cap on the end of one of the β-subunit helices. Arrow, bilin-binding site. c Internal duplication of GtcpeA10. The mature sequence was split into N- and C-terminal halves and aligned to show sequence relatedness. Both halves a bilin-binding site. Peptides identified by mass spectrometry are underlined. Parenthesis, removed N-terminal Met; arrow, bilin-binding site