Abstract
Tooth agenesis is one of the most common developmental anomalies affecting function and esthetics. The paired-domain transcription factor, Pax9, is critical for patterning and morphogenesis of tooth and taste buds. Mutations of PAX9 have been identified in patients with tooth agenesis. Despite significant progress in the genetics of tooth agenesis, many gaps in knowledge exist in refining the genotype-phenotype correlation between PAX9 and tooth agenesis. In the present study, we complete genetic and phenotypic characterization of multiplex Chinese families with nonsyndromic (NS) tooth agenesis. Direct sequencing of polymerase chain reaction products revealed 9 novel (c.140G>C, c.167T>A, c.332G>C, c.194C>A, c.271A>T, c.146delC, c.185_189dup, c.256_262dup, and c.592delG) and 2 known heterozygous mutations in the PAX9 gene among 120 probands. Subsequently, pedigrees were extended, and we confirmed that the mutations co-segregated with the tooth agenesis phenotype (with exception of families in which DNA analysis was not available). In 1 family (n = 6), 2 individuals harbored both the PAX9 c.592delG mutation and a heterozygous missense mutation (c.739C>T) in the MSX1 gene. Clinical characterization of families segregating a PAX9 mutation reveal that all affected individuals were missing the mandibular second molar and their maxillary central incisors are most susceptible to microdontia. A significant reduction of bitter taste perception was documented in individuals harboring PAX9 mutations (n = 3). Functional studies revealed that PAX9 haploinsufficiency or a loss of function of the PAX9 protein underlies tooth agenesis.
Keywords: dental agenesis, oligodontia, phenotypic analysis, craniofacial anomalies, craniofacial genetics, functional studies
Introduction
Nonsyndromic (NS) tooth agenesis is one of the most common congenital disorders, with a prevalence rate from 2.2% to 10.1% in the general population, when third molars are excluded (Polder et al. 2004; Zhang, Liu, et al. 2015). Based on the number of missing teeth, tooth agenesis can be classified into hypodontia (less than 6 missing teeth), oligodontia (6 or more missing teeth), and anodontia (complete agenesis of permanent dentition). Multiple lines of evidence demonstrate that environmental and genetic factors contribute to the development of tooth agenesis (Nieminen 2009; Yin and Bian 2015). Compared with hypodontia, oligodontia is more severe and rare (0.14% to 0.30% in the population) (Dhanrajani 2002), and has a stronger genetic basis (Liu et al. 2013); more than half of oligodontia patients carry gene mutations (van den Boogaard et al. 2012; Song et al. 2014). To date, mutations in at least 10 genes, including MSX1, PAX9, AXIN2, EDA, WNT10A, EDARADD, KRT17, LRP6, and WNT10B, have been identified in patients with tooth agenesis (Song et al. 2009; Stockton et al. 2000; Ruf et al. 2013; Wong, Liu, Bai, et al. 2014; Wong, Liu, Han, et al. 2014; Massink et al. 2015; Yu et al. 2016). Implicitly, the understanding of genes involved in tooth agenesis necessitates an understanding of genes responsible for normal tooth development. Over 300 transcription factors, signaling molecules, receptors, and so on (http://bite-it.helsinki.fi/) are known to be expressed during tooth development in the spatial and temporal pattern (Jernvall and Thesleff 2000), which complicates our ability to understand the specific role that each plays in odontogenesis. Therefore, despite the significant advances in dental developmental genetics over the past 2 decades (Yin and Bian 2015), we still do not fully understand all of the sufficient or necessary molecular pathways that result in the dentition or, more specifically, how each gene or alteration leads to a specific phenotype.
Among the many genes identified in normal tooth development and, by extension, tooth agenesis, the PAX9 gene provides a unique opportunity to interrogate the genotype-phenotype correlation in human tooth agenesis. Indeed, this critical transcription factor is expressed in the presumptive dental mesenchyme to activate odontogenic signals and to initiate tooth development; evidence from Pax9-deficient mice illustrated an arrest of tooth development at the early bud stage (Peters et al. 1998). In addition to the function of dental organogenesis, a recent study indicates that Pax9 is crucial for the differentiation and formation of taste buds of the tongue, as Pax9 knockout mice show the arrested development of circumvallate papillae and foliate papillae (Kist et al. 2014).
Despite advances in molecular genetics and animal studies discussed above, important questions remain unanswered regarding the genetic etiology and phenotypic variability of NS tooth agenesis (Frazier-Bowers and Vora 2017). In this report, we seek to enhance the knowledge base of phenotypic variation within the tooth agenesis spectrum. Our study takes advantage of multiplex families with a shared racial and genetic background that segregate mutations in the PAX9 gene. Accordingly, mutational analysis in a cohort of 120 unrelated patients of tooth agenesis identified 11 PAX9 mutations, including 9 novel mutations. In this study, we report novel genotype-phenotype correlations that include pattern of tooth agenesis, size and shape of teeth, and phenotypic characterization of another ectodermal derivative, taste buds. Taken together, these results add to the knowledge base, primarily how one gene within a specific molecular pathway links to tooth agenesis and how different genetic alterations lead to subtle differences in tooth agenesis phenotypes.
Materials and Methods
Subjects
A cohort of 120 unrelated patients with tooth agenesis (tooth agenesis number ≥6, excluding third molars) was recruited to participate in this study by referral from the Department of Prosthodontics in Peking University Hospital of Stomatology (PKUSS) and the Department of Prosthodontics, Beijing Stomatological Hospital from 2008 to 2015. These patients denied that their missing permanent teeth were due to extraction and injuries. Phenotypic characterization for all patients included intraoral examination and panoramic radiographs to verify the number and pattern of missing teeth. Informed consent was obtained to complete genetic testing and clinical photographs for each patient over 18 y of age, and an assent and parental consent were obtained for minors (under 18 y). Healthy control patients (n = 100) were also recruited (18 to 40 y of age) from the orthodontic clinic. This included the same clinical photos and panoramic radiographs taken for affected subjects. This study was approved by institutional review board of PKUSS.
Mutation Analysis
Genomic DNA was extracted from peripheral blood lymphocytes as previous described (Wong, Liu, Bai, et al. 2014). Four exons and intron-exon boundaries of the PAX9 gene and 2 exons and intron-exon boundaries of MSX1 were amplified by polymerase chain reaction (PCR). PCR products were sent to Sangon Biotech Company for direct sequencing. PCR sequences identified with a mutation were cloned into pGEM-T easy vectors (Tiangen), followed by sequencing of isolated clones.
Taste Function Evaluation
Four tastants were used to evaluate the taste sensitivity for available affected subjects who carried a PAX9 mutation (n = 3) and 10 control subjects at the following concentrations: 0.4, 0.2, 0.1, and 0.05 g/mL sucrose (sweet); 0.3, 0.165, 0.09, and 0.05 g/mL citric acid (sour); 0.25, 0.1, 0.04, and 0.016 g/mL sodium chloride (salty); 0.006, 0.0024, 0.0009, and 0.0004 g/mL quinine hydrochloride (bitter) (Mueller et al. 2003). Briefly, filter paper strips were impregnated with 4 tastants with different concentrations and placed on the left and then right side of anterior third of the extended tongue (Landis et al. 2009). Before testing, the mouth was rinsed with water. Individuals were asked to identify the taste, and the taste score was obtained. Each taste was scored on a scale of 0 to 4 from one side of tongue, with bigger values indicating more sensitivity in this taste. Therefore, a maximum of 32 points would be obtained from both sides of the tongue.
Construction of PAX9 Expression Vectors and Site-Directed Mutagenesis
The human PAX9 complementary DNA (cDNA) (accession number: NM_006194; Origene) was subcloned into the pCMV-C-Myc expression vector (Beyotime) between 5′-BamHI and 3′-EcoRI sites to generate the pCMV-PAX9-Myc wild-type plasmid. Site-directed mutagenesis by the QuickChange Lightning Site-Directed Mutagenesis Kit (Agilent) was performed to generate 4 missense mutants (R26W, R47P, I56N, and A108P) from the wild-type construct. For the frameshift mutants (146delC, 185_189dup, 256_262dup, and 592delC) generation, we employed a 2-step process: 1) using a site-directed mutagenesis system to insert or delete nucleotides, causing a shift of the reading frame, and 2) using a 3′-EcoRI primer designed prior to the premature stop codon and a 5′-BamHI primer at the start codon to amplify truncated fragments by PCR, and then the fragments were subcloned into the expression vector prior to the Myc-epitope. Two nonsense mutant (S56* and K91*) vectors were constructed in the same manner by designing primers with enzyme restriction sites. All mutant constructs were verified by sequencing.
Expression of Wild-Type and Mutant Plasmids
COS-7 cells were maintained in Dulbecco’s modified Eagle’s medium with 10% fetal bovine serum (Gibco). Lipofectamine (Invitrogen) was used for transient transfection of wild-type and mutant plasmids. Forty-eight hours after transfection, protein was harvested. Whole-cell lysates (30 µg) were separated by 12% polyacrylamide gel and were then electroblotted to an Immobilon-P membrane (Millipore). Primary antibodies against β-actin, PAX9, and Myc-tag from Cell Signaling (Danvers) were used for overnight incubation. Blotting was performed as previously described (Wong, Liu, Han, et al. 2014).
Real-Time PCR and Messenger RNA Stability Studies
Twelve hours after transfection, total RNA was isolated by the RNeasy plus mini kit (Qiagen) and was reverse transcripted using the Qiagen Reverse Transcription Kit. We designed a specific pair of primers, PAX9-F: 5′-AACCAGCTGGGAGGAGTGTT-3′ and PAX9-R: 5′-TGATGTCACACGGTCGGATG-3′, located at the N-terminus of the paired box domain as they can recognize the wild-type and all mutant messenger RNAs (mRNAs). SYBR green–based quantitative reverse transcription (RT)–PCR was performed using the Applied Biosystems 7900 Real Time PCR System. The expression of PAX9 was normalized by eukaryotic 18S ribosomal RNA (rRNA). The mRNA stability assessment was modified from a previous study (Suda et al. 2011). Briefly, 12 h after transfection, cells were treated with 10 µg/mL actinomycin D (Gibco). After 4- and 8-h treatments, total RNA was harvested for reverse transcription. The mRNA stability was presented as the percentage of remaining PAX9 mRNA, normalized by 18rRNA, after actinomycin D treatment.
Immunofluorescence
Forty-eight hours after transfection, cells were fixed with 95% ethanol and permeablilized by 0.1% Triton-X. After blocking with 10% goat serum, anti-Myc tag primary antibody (1:200) was incubated at 4°C overnight. Tetramethyl rhodamine isocyanate (TRITC)–conjugated goat anti-rabbit antibody (Origene) was incubated at room temperature for 1 h. Fluorescein isothiocyanate (FITC)–conjugated phalloidin (Sigma) was used for staining cytoskeleton. Cells were sealed with mounting medium containing DAPI. Images were taken using the confocal microscope LSM 510 Meta (Zeiss) with a ×40/1.00 numerical aperture oil objective lens.
Electrophoretic Mobility Shift Assay
Paired domain high-affinity binding probe CD19-2(A-ins) (Czerny and Busslinger 1995) was synthesized and its 5′ end was labeled with biotin. Electrophoretic mobility shift assay (EMSA) was performed using the LightShift Chemiluminescent EMSA Kit (Thermo Scientific) according to the manufacturer’s instructions. Forty-eight hours after transfection of the expression plasmids, 5 µg of nuclear extracts was incubated with annealed oligonucleotides at room temperature for 20 min. The protein-DNA complexes were electrophoresed on a 6% nondenaturing polyacrylamide gel in 0.5× Tris borate EDTA, transferred to a nylon membrane, and imaged by FUSION-FX Chemiluminescence System (Vilber Lourmat).
Luciferase Reporter Assay
PAX9 expression vectors were cotransfected with the reporter plasmid containing human BMP4 promoter (p.2.8 BMP4-Luc) as previously described (Kawai and Sugiura 2001; Liang et al. 2012); the phRL-TK plasmid was used as an internal control. Forty-eight hours after transfection, the cell extracts were assayed using the Dual-Luciferase Reporter Assay System (Promega) according to the manufacturer’s instructions. Firefly luciferase activity was normalized based on Renilla luciferase activity.
Statistical Analysis
Statistical analyses were performed with SAS 9.2 (SAS Institute). The χ2 test was used to compare the frequencies. To compare taste scores between groups, the Mann-Whitney test was used. Analysis of variance (ANOVA) test or Student’s t test was used for mRNA expression and stability studies, as well as for the luciferase reporter assay. A P value <0.05 was considered statistically significant.
Results
Mutational Analysis and Clinical Findings
Mutation screening of PAX9 and MSX1 of 120 nonconsanguineous patients with tooth agenesis revealed 11 unrelated individuals (9.12%) with distinct PAX9 mutations (Table and Appendix Fig. 1). All patients had normal facial appearance, hair, skin, and nails and thus were diagnosed with nonsyndromic tooth agenesis. The 11 mutations included 4 missense, 2 nonsense, and 5 frameshift mutations, all located at exon 2 of the PAX9 gene (Table). Nine of them (c.140G>C, c.167T>A, c.332G>C, c.194C>A, c.271A>T, c.146delC, c.185_189dup, c.256_262dup, c.592delG) are novel mutations. Pedigree analysis by visual inspection suggested that the tooth agenesis trait is autosomal dominant in 10 of 11 families (Appendix Fig. 2), and we have confirmed the mutant allele was inherited through either paternal or maternal transmission in a complete penetrance (Table). The proband (II: 1) of family 9 (PAX9 c.146delC) does not have a family history of tooth agenesis, but we confirmed that this is a de novo mutation. Mutations identified in affected individuals were not seen in the healthy controls (n = 100) and ExAC database, confirming that they were absent in the general population.
Table.
Summary of PAX9 Mutations in Patients with Tooth Agenesis.
| Family No./Proband No. | Sex/Age, y | Exon | Nucleotide Change | Protein Change | Type of Mutation | Hereditary |
|---|---|---|---|---|---|---|
| H40/III:4 | M/17 | 2 | c.76C>T | p.Arg26Trp | Missense | Paternal |
| H42/III:1 | M/16 | 2 | c.140G>C | p.Arg47Pro | Missense | Paternal |
| 9/II:1 | F/18 | 2 | c.146delC | p.Ser49Cysfs*36 | Frameshift | De novo |
| HY3/II:1 | F/26 | 2 | c.167T>A | p.Ile56Asn | Missense | Not test |
| H37/III:1 | M/8 | 2 | c.185_189dup | p.Gly64Argfs*23 | Frameshift | Paternal |
| 69/II:1 | M/27 | 2 | c.194C>A | p.Ser65* | Nonsense | Paternal |
| HY13/III:1 | F/18 | 2 | c.218dupG | p.Ser74Glnfs*243 | Frameshift | Paternal |
| HY40/II:5 | F/46 | 2 | c.256_262dup | p.Arg88Profs*231 | Frameshift | Not test |
| H46/III:2 | F/23 | 2 | c.322G>C | p.Ala108Pro | Missense | Paternal |
| W37/II:1 | M/17 | 2 | c.271A>T | p.Lys91* | Nonsense | Not test |
| HY54/III:6 | F/23 | 2 | c.592delG | p.Val198Serfs*14 | Frameshift | Maternal |
F, female; M, male.
The proband (III: 6) of family HY54 carrying the c.592delG mutation in PAX9 also segregated a MSX1 missense mutation (c.739C>T) (Appendix Fig. 3B), with both mutations inherited from her mother (II: 7), who also presented with oligodontia (Appendix Fig. 3A). The PAX9 c.592delG mutation caused a frameshift and premature termination of PAX9 protein (Appendix Fig. 3C). The MSX1 c.739C>T mutation is predicted to be a functional mutation (probability = 0.99/1) by MutationTaster as the affected residue is evolutionarily conserved (Appendix Fig. 3D). The identification of the MSX1 mutation, although predicted as functional, does not likely confer a significant contribution to the tooth agenesis phenotype noted here (Appendix Fig. 3A).
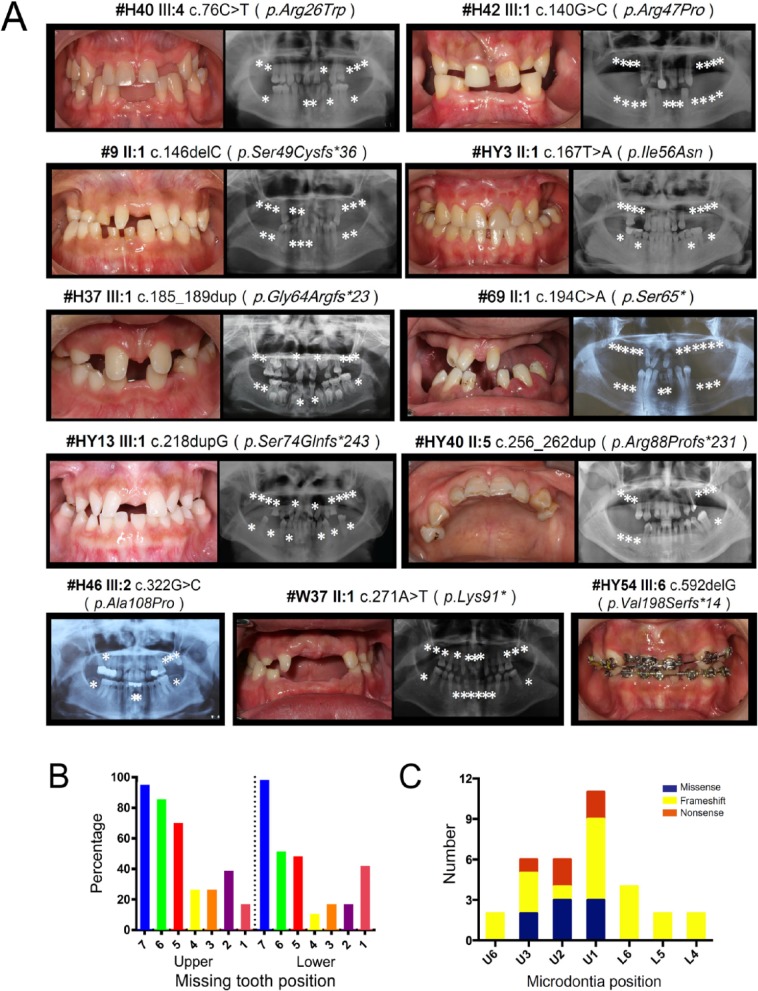
Analysis of the tooth agenesis pattern in affected individuals with PAX9 mutations (Fig. 1 and Appendix Table 1) revealed that the most common pattern of missing teeth in patients with PAX9 mutations (n = 16 patients) is as follows: lower second molars (L7), with a 100% affected rate, and least affected lower first bicuspids (L4), with a 12.5% missing rate (Fig. 1B). The prevalence of missing teeth in patients with a PAX9 nonsense mutation (66.1%) was higher than those with a PAX9 frameshift mutation (49.6%) or a missense mutation (40.5%) (P = 0.004) (Appendix Fig. 4, Appendix Table 2). The upper central incisor (U1) was most susceptible to microdontia (Fig. 1C). The prevalence of microdontia in the missense mutation group (4.76%) was less than the frameshift mutation group (9.8%) and nonsense (9.8%) mutation group, and the difference was statistically significant (P < 0.001) (Appendix Table 2).
Figure 1.
Clinical characteristics of probands with a PAX9 mutation. (A) Intraoral photos and panoramic radiographs of probands with distinct PAX9 mutations. Asterisks represent the position of missing teeth. (B) The prevalence of missing teeth in all PAX9 mutation patients (n = 16). L, lower; U, upper. (C) The prevalence of microdontia in all patients with a PAX9 mutation.
We further assessed the gustatory function in available individuals with PAX9 variants (n = 3) by taste testing. A significant reduction of bitter taste perception was observed in the PAX9 mutant patients relative to normal controls (P = 0.024), but no significant differences were found in sweet, salty, sour, and total taste scores between the 2 groups (Appendix Table 3).
Effects of Mutations on PAX9 Function
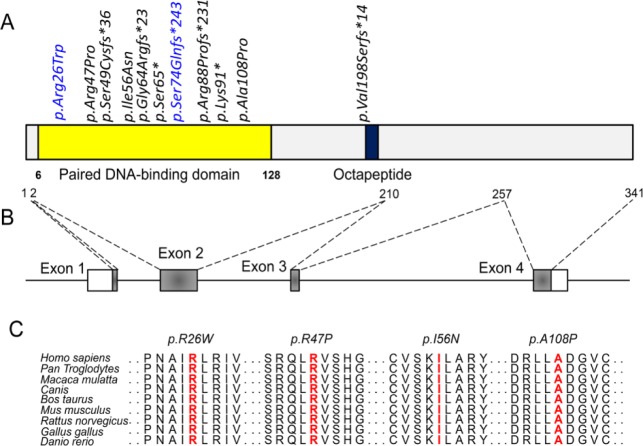
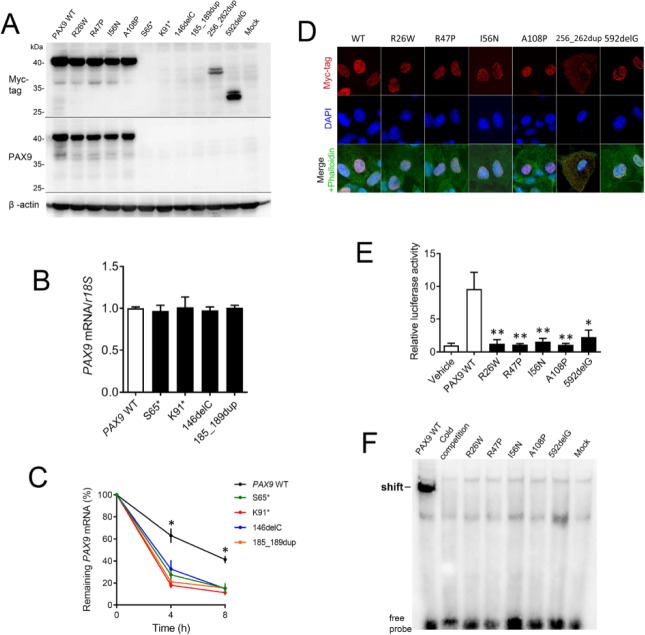
Functional studies were completed for all mutants affected by novel PAX9 mutations. First, a conservation analysis revealed that 4 missense mutations—Arg26, Arg47, Ile56, and Ala108—all located at the paired DNA-binding domain are highly conserved during evolution (Fig. 2). To confirm that the mutations affect PAX9 function and thus contribute to the pathogenesis of tooth agenesis, in vitro studies of mutant PAX9 proteins were performed using plasmids containing 9 novel PAX9 mutations. Western blot (Fig. 3A) demonstrated that all mutant proteins caused by missense mutations and 2 truncated proteins (256_262dup and 592delG) caused by frameshift mutations were expressed in vitro. However, mutant proteins caused by nonsense mutations (S56* and K91*) and frameshift mutations (145delC and 185_189dup) were repeatedly undetectable. For those mutants that failed to express proteins in vitro, we further examined their mRNA expression levels. Although baseline expression levels of truncated mRNAs did not differ from the wild-type PAX9 (P > 0.05), after actinomycin D treatment, the percentage of remaining mutant mRNAs was significantly less than that of wild-type mRNA (P < 0.05). These results suggest that these truncated mRNAs were less stable and were more likely to undergo decay compared to the normal PAX9.
Figure 2.
Location and conservation analysis of tooth agenesis associated PAX9 mutations. (A) Distribution of mutations identified in patients with tooth agenesis in the PAX9 protein. Reported mutations are labeled in blue. Black indicates novel PAX9 mutations. (B) Schematic diagram of the PAX9 gene. (C) Conservation analysis of affected amino acids in the PAX9 protein among 9 different vertebrate species.
Figure 3.
Functional studies of mutant PAX9 proteins. (A) Expression of wild-type and mutant PAX9 proteins detected by Western blot. (B) Relative messenger RNA (mRNA) expression levels of mutant PAX9 at 12 h posttransfection. (C) After treatment with actinomycin D, PAX9 mRNAs underwent decay. The mRNA stability is presented by the percentage of remaining mRNAs at 4 and 8 h posttreatment. *P < 0.05. (D) Subcellular localization of wild-type and mutant PAX9 proteins assessed by immunofluorescence. (E) The transcriptional activation abilities of PAX9 mutants on the BMP4 promoter assessed by luciferase reporter assay. **P < 0.01. (F) Binding of PAX9 mutants to the paired domain consensus sequence CD19-2 (A-ins) assessed by an electrophoretic mobility shift assay.
We examined whether the mutations affect the nuclear localization of expressed protein. All missense mutants and the 592delG frameshift mutant localized in nuclei of transient transfected COS-7 cells resembling the localization of normal PAX9 (Fig. 3B), whereas another frameshift mutant (256_262dup) localized at the entire cytoplasm. We next sought to understand whether the mutations affect the transcriptional activation of the BMP4 promoter (a downstream target of PAX9) and interfere with DNA binding. In luciferase experiments using a PAX9 response element from the BMP4 gene, all mutants (R26W, R47P, I56N, A108P, and 592delG) failed to activate the reporter (Fig. 3E). The gel shift analysis further demonstrated that these mutants all lost their ability to bind the DNA consensus Pax sequence (Fig. 3F).
Discussion
To date, about 30 mutations have been reported in the PAX9 gene associated with tooth agenesis (Liang et al. 2016). All but 2 of these (c.1A>G, c2T>G) are located in exon 1 (Klein et al. 2005; Liang et al. 2016), with 1 (c.792_793insC) occurring in exon 4 (Frazier-Bowers et al. 2002), and 27 PAX9 mutations are located in exon 2 (Liang et al. 2016). All of the PAX9 mutations in our study (n = 11) distributed within exon 2 (Fig. 2), confirming that this exon is the germline mutation hotspot. Exon 2 comprises 61.11% of the coding sequence of PAX9, encoding the evolutionarily conserved DNA-binding domain, the paired box domain (Neubuser et al. 1995). We found that 10 PAX9 mutations occurred in the DNA-binding domain (Fig. 2A). These results support other findings that the high evolutionary constraint region is closely associated with clusters of mutational hotspots (Walker et al. 1999) and suggest that evolutionary conservation is a good surrogate marker for mutational hotspots in the PAX9 gene.
In this study, we corroborate the previous molecular studies of PAX9 mutation contribution to tooth agenesis but also provide a higher resolution of phenotypic analysis. It has already been shown that Pax9 is critical for molar development; hence, patients carrying PAX9 mutations commonly present the agenesis of molars (Kapadia et al. 2006). In our study, we highlight a novel finding that the lower second molar is most commonly affected, with a 100% missing rate. Lower first bicuspids are the least affected, with a 12.5% missing rate, suggesting that PAX9 is less crucial for its development compared with molars. We also found in our sample that the upper central incisors are most susceptible to cone-shape tooth and posit that this underscores the importance of normal PAX9 in regulating the tooth shape of maxillary central incisors.
The observation of digenic mutations in MSX1 and PAX9 in our study presents with a more severe phenotype (missing number n = 15 or 17) compared to the family member with a PAX9 monogenic mutation (missing number n = 12). However, in previous reports, the digenic mutations appear to be inconclusive relative to genotype-phenotype correlation in tooth agenesis (Arte et al. 2013; He et al. 2013; Kantaputra et al. 2015; Zhang, Wong, et al. 2015). One can speculate that since Pax9 has been shown to interact with Msx1 and synergistically activate the expression of downstream dental organogenesis genes (Ogawa et al. 2006), mutation in both genes might serve to abolish the synergistic effect and thus results in a more severe tooth agenesis phenotype.
Although Pax9 is widely expressed in forelimbs, pharyngeal pouches, and many craniofacial sites, including mandibularies, the first branchial arch, nasal mesenchyme, and the tongue, during embryogenesis (Neubuser et al. 1995), dentition is usually exclusively affected in patients with PAX9 loss-of-function mutations (Liang et al. 2016). In a previous study, 1 family with a PAX9 frameshift mutation had hair problems in addition to dental agenesis (Mostowska et al. 2013). Therefore, we sought to explore the full range of a PAX9 mutation on the broader phenotype and assessed another ectodermal derivative (i.e., not just limited to teeth), taste buds. Our finding documenting a significant decrease in bitter perception suggests that PAX9 mutations might also affect the development and function of circumvallate and foliate papillae, both of which comprise the G protein–coupled taste receptor TAS2R that detects bitterness (Chandrashekar et al. 2000). However, the sample size of available patients was relatively small (n = 3), and thus these findings need to be further confirmed.
Overall, the results of this study reveal that the severity of tooth loss is associated with the type of mutation, which results in different degrees of loss of function in PAX9. Individuals with a nonsense mutation have a more severe tooth agenesis phenotype compared with those with a frameshift or a missense mutation in PAX9. Our in vitro studies showed that these nonsense mutants were undetectable and might be triggered by an RNA surveillance system (e.g., nonsense-mediated mRNA decay to prevent the production of truncated proteins) (Akimitsu 2008). Indeed, in vitro results demonstrate that the truncated mRNAs were less stable and more likely to undergo decay. The missense mutants were expressed in vitro and had a normal subcellular localization, but they failed to activate the transcription of BMP4 due to the defective DNA binding. Correspondingly, the individuals with missense mutations had a mild phenotype with the least rate of missing teeth compared to those with frameshift or nonsense mutations. The frameshift mutations resulted in different rates of loss of function in PAX9, and thus its tooth missing rate is higher than that of the missense mutation but lower than that of the nonsense mutation. Therefore, our study supports previous findings that truncating mutations have a more deleterious effect compared to missense mutations (Liang et al. 2016).
In summary, we have found 9 novel PAX9 mutations in patients with tooth agenesis that greatly expand the mutation spectrum. We further performed detailed clinical phenotype characterization that refines and broadens the phenotype spectrum of PAX9-associated tooth agenesis. We conclude that loss of function of the PAX9 mutations causes tooth agenesis among these patients.
Author Contributions
S.-W. Wong, contributed to conception, design, data acquisition, analysis, and interpretation, drafted and critically revised the manuscript; D. Han, contributed to conception and data acquisition, critically revised the manuscript; H. Zhang, Y. Liu, X. Zhang, Y. Wang, N. Zhao, L. Zeng, B. Bai, contributed to data acquisition, critically revised the manuscript; M.Z. Miao, contributed to data analysis, critically revised the manuscript; Y.-X. Wang, contributed to conception and data interpretation, critically revised the manuscript; H. Liu, S.A. Frazier-Bowers, H. Feng, contributed to conception, design, data analysis, and interpretation, drafted and critically revised the manuscript. All authors gave final approval and agree to be accountable for all aspects of the work.
Supplementary Material
Acknowledgments
We are grateful to the families with tooth agenesis for their participation in this study.
Footnotes
A supplemental appendix to this article is available online.
This work was supported by Natural Science Foundation of China grant (81271121 to H. Feng, 81600851 to H. Liu, 81600846 to Y. Liu), Capital Medical Developing Foundation (2007-1005 to H. Feng and B. Bai) and the National Institute of Dental and Craniofacial Research (R90DE022527 to S.-W. Wong).
The authors declare no potential conflicts of interest with respect to the authorship and/or publication of this article.
Web Resources: Ensembl: http://www.ensembl.org/
ExAC Browser: http://exac.broadinstitute.org/
MutationTaster: http://www.mutationtaster.org/
Mutalyzer: https://www.mutalyzer.nl/
OMIM: http://www.omim.org
References
- Akimitsu N. 2008. Messenger RNA surveillance systems monitoring proper translation termination. J Biochem. 143(1):1–8. [DOI] [PubMed] [Google Scholar]
- Arte S, Parmanen S, Pirinen S, Alaluusua S, Nieminen P. 2013. Candidate gene analysis of tooth agenesis identifies novel mutations in six genes and suggests significant role for WNT and EDA signaling and allele combinations. PLoS One. 8(8):e73705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandrashekar J, Mueller KL, Hoon MA, Adler E, Feng L, Guo W, Zuker CS, Ryba NJ. 2000. T2rs function as bitter taste receptors. Cell. 100(6):703–711. [DOI] [PubMed] [Google Scholar]
- Czerny T, Busslinger M. 1995. DNA-binding and transactivation properties of Pax-6: three amino acids in the paired domain are responsible for the different sequence recognition of Pax-6 and BSAP (Pax-5). Mol Cell Biol. 15(5):2858–2871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhanrajani PJ. 2002. Hypodontia: etiology, clinical features, and management. Quintessence Int. 33(4):294–302. [PubMed] [Google Scholar]
- Frazier-Bowers SA, Guo DC, Cavender A, Xue L, Evans B, King T, Milewicz D, D’Souza RN. 2002. A novel mutation in human Pax9 causes molar oligodontia. J Dent Res. 81(2):129–133. [PubMed] [Google Scholar]
- Frazier-Bowers SA, Vora SR. 2017. Genetic disorders of dental development: tales from the bony crypt. Curr Osteoporos Rep. 15(1):9–17. [DOI] [PubMed] [Google Scholar]
- He H, Han D, Feng H, Qu H, Song S, Bai B, Zhang Z. 2013. Involvement of and interaction between WNT10A and EDA mutations in tooth agenesis cases in the Chinese population. PLoS One. 8(11):e80393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jernvall J, Thesleff I. 2000. Reiterative signaling and patterning during mammalian tooth morphogenesis. Mech Dev. 92(1):19–29. [DOI] [PubMed] [Google Scholar]
- Kantaputra PN, Kaewgahya M, Hatsadaloi A, Vogel P, Kawasaki K, Ohazama A, Ketudat Cairns JR. 2015. Gremlin 2 mutations and dental anomalies. J Dent Res. 94(12):1646–1652. [DOI] [PubMed] [Google Scholar]
- Kapadia H, Frazier-Bowers S, Ogawa T, D’Souza RN. 2006. Molecular characterization of a novel Pax9 missense mutation causing posterior tooth agenesis. Eur J Hum Genet. 14(4):403–409. [DOI] [PubMed] [Google Scholar]
- Kawai S, Sugiura T. 2001. Characterization of human bone morphogenetic protein (BMP)-4 and -7 gene promoters: activation of BMP promoters by GLI, a sonic hedgehog mediator. Bone. 29(1):54–61. [DOI] [PubMed] [Google Scholar]
- Kist R, Watson M, Crosier M, Robinson M, Fuchs J, Reichelt J, Peters H. 2014. The formation of endoderm-derived taste sensory organs requires a pax9-dependent expansion of embryonic taste bud progenitor cells. PLoS Genet. 10(10):e1004709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein ML, Nieminen P, Lammi L, Niebuhr E, Kreiborg S. 2005. Novel mutation of the initiation codon of Pax9 causes oligodontia. J Dent Res. 84(1):43–47. [DOI] [PubMed] [Google Scholar]
- Landis BN, Welge-Luessen A, Bramerson A, Bende M, Mueller CA, Nordin S, Hummel T. 2009. “Taste strips”—a rapid, lateralized, gustatory bedside identification test based on impregnated filter papers. J Neurol. 256(2):242–248. [DOI] [PubMed] [Google Scholar]
- Liang J, Qin C, Yue H, He H, Bian Z. 2016. A novel initiation codon mutation of Pax9 in a family with oligodontia. Arch Oral Biol. 61:144–148. [DOI] [PubMed] [Google Scholar]
- Liang J, Song G, Li Q, Bian Z. 2012. Novel missense mutations in Pax9 causing oligodontia. Arch Oral Biol. 57(6):784–789. [DOI] [PubMed] [Google Scholar]
- Liu H, Han D, Wong S, Nan X, Zhao H, Feng H. 2013. Rs929387 of GLI3 is involved in tooth agenesis in Chinese Han population. PLoS One. 8(11):e80860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massink MP, Creton MA, Spanevello F, Fennis WM, Cune MS, Savelberg SM, Nijman IJ, Maurice MM, van den Boogaard MJ, van Haaften G. 2015. Loss-of-function mutations in the WNT co-receptor LRP6 cause autosomal-dominant oligodontia. Am J Hum Genet. 97(4):621–626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mostowska A, Zadurska M, Rakowska A, Lianeri M, Jagodzinski PP. 2013. Novel Pax9 mutation associated with syndromic tooth agenesis. Eur J Oral Sci. 121(5):403–411. [DOI] [PubMed] [Google Scholar]
- Mueller C, Kallert S, Renner B, Stiassny K, Temmel AF, Hummel T, Kobal G. 2003. Quantitative assessment of gustatory function in a clinical context using impregnated “taste strips.” Rhinology. 41(1):2–6. [PubMed] [Google Scholar]
- Neubuser A, Koseki H, Balling R. 1995. Characterization and developmental expression of Pax9, a paired-box-containing gene related to Pax1. Dev Biol. 170(2):701–716. [DOI] [PubMed] [Google Scholar]
- Nieminen P. 2009. Genetic basis of tooth agenesis. J Exp Zool B Mol Dev Evol. 312(4):320–342. [DOI] [PubMed] [Google Scholar]
- Ogawa T, Kapadia H, Feng JQ, Raghow R, Peters H, D’Souza RN. 2006. Functional consequences of interactions between Pax9 and Msx1 genes in normal and abnormal tooth development. J Biol Chem. 281(27):18363–18369. [DOI] [PubMed] [Google Scholar]
- Peters H, Neubuser A, Kratochwil K, Balling R. 1998. Pax9-deficient mice lack pharyngeal pouch derivatives and teeth and exhibit craniofacial and limb abnormalities. Genes Dev. 12(17):2735–2747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polder BJ, Van’t Hof MA, Van der Linden FP, Kuijpers-Jagtman AM. 2004. A meta-analysis of the prevalence of dental agenesis of permanent teeth. Community Dent Oral Epidemiol. 32(3):217–226. [DOI] [PubMed] [Google Scholar]
- Ruf S, Klimas D, Honemann M, Jabir S. 2013. Genetic background of nonsyndromic oligodontia: a systematic review and meta-analysis. J Orofac Orthop. 74(4):295–308. [DOI] [PubMed] [Google Scholar]
- Song S, Han D, Qu H, Gong Y, Wu H, Zhang X, Zhong N, Feng H. 2009. EDA gene mutations underlie non-syndromic oligodontia. J Dent Res. 88(2):126–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song S, Zhao R, He H, Zhang J, Feng H, Lin L. 2014. Wnt10a variants are associated with non-syndromic tooth agenesis in the general population. Hum Genet. 133(1):117–124. [DOI] [PubMed] [Google Scholar]
- Stockton DW, Das P, Goldenberg M, D’Souza RN, Patel PI. 2000. Mutation of Pax9 is associated with oligodontia. Nat Genet. 24(1):18–19. [DOI] [PubMed] [Google Scholar]
- Suda N, Ogawa T, Kojima T, Saito C, Moriyama K. 2011. Non-syndromic oligodontia with a novel mutation of Pax9. J Dent Res. 90(3):382–386. [DOI] [PubMed] [Google Scholar]
- van den Boogaard MJ, Creton M, Bronkhorst Y, van der Hout A, Hennekam E, Lindhout D, Cune M, Ploos van Amstel HK. 2012. Mutations in WNT10A are present in more than half of isolated hypodontia cases. J Med Genet. 49(5):327–331. [DOI] [PubMed] [Google Scholar]
- Walker DR, Bond JP, Tarone RE, Harris CC, Makalowski W, Boguski MS, Greenblatt MS. 1999. Evolutionary conservation and somatic mutation hotspot maps of p53: correlation with p53 protein structural and functional features. Oncogene. 18(1):211–218. [DOI] [PubMed] [Google Scholar]
- Wong S, Liu H, Bai B, Chang H, Zhao H, Wang Y, Han D, Feng H. 2014. Novel missense mutations in the AXIN2 gene associated with non-syndromic oligodontia. Arch Oral Biol. 59(3):349–353. [DOI] [PubMed] [Google Scholar]
- Wong SW, Liu HC, Han D, Chang HG, Zhao HS, Wang YX, Feng HL. 2014. A novel non-stop mutation in MSX1 causing autosomal dominant non-syndromic oligodontia. Mutagenesis. 29(5):319–323. [DOI] [PubMed] [Google Scholar]
- Yin W, Bian Z. 2015. The gene network underlying hypodontia. J Dent Res. 94(7):878–885. [DOI] [PubMed] [Google Scholar]
- Yu P, Yang W, Han D, Wang X, Guo S, Li J, Li F, Zhang X, Wong SW, Bai B, et al. 2016. Mutations in WNT10B are identified in individuals with oligodontia. Am J Hum Genet. 99(1):195–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Liu HC, Lyu X, Shen GH, Deng XX, Li WR, Zhang XX, Feng HL. 2015. Prevalence of tooth agenesis in adolescent Chinese populations with or without orthodontics. Chin J Dent Res. 18(1):59–65. [PubMed] [Google Scholar]
- Zhang XX, Wong SW, Han D, Feng HL. 2015. Simultaneous occurrence of an autosomal dominant inherited MSX1 mutation and an X-linked recessive inherited EDA mutation in one Chinese family with non-syndromic oligodontia. Chin J Dent Res. 18(4):229–234. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.