Table 3.
Differential expression and σA promoter region of genes with known or predicted DtxR binding site. Based on the differential expression values the genes are classified into three groups: repressed, activated and not differentially expressed. In case no TSS could be assigned to the gene only the translation start codon is shown. In case a TSS could be assigned, but no −10 motif could be identified the start codon is preceded with a dotted line. Counts, normalized read counts; LFC, log2(fold change); Start, start codon. The asterisks mark genes with an experimentally shown DtxR regulation
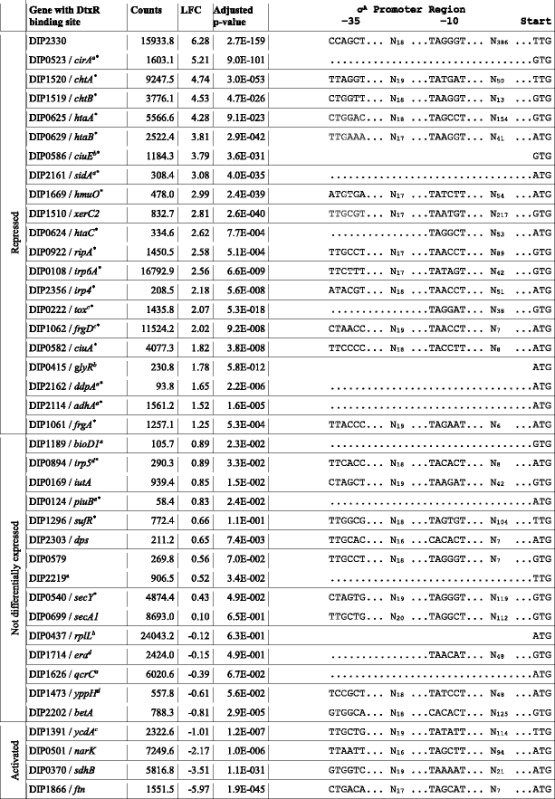
aNo TSS detected, hence no promoter motif predictable
bGene is part of a primary operon and lacks own TSS
cMultiple TSS detected and only TSS closest to DtxR binding site shown. In case of the tox gene the −10 motif of the TSS (TSS 2) closest to the start codon is shown
dGene is the first one in a sub-operon and therefore has own TSS
