Fig. 1.
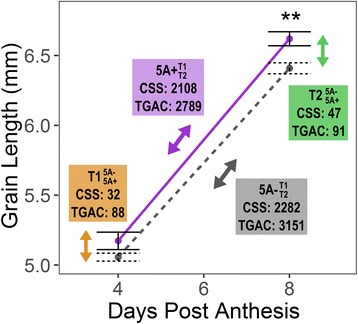
Differentially expressed genes between 5A NILs across time. RNA-seq was carried out on whole grain RNA samples taken in 4 different conditions from NILs grown in 2014 [29]: 5A- (short grains) and 5A+ (long grains) NILs at 4 days post anthesis (dpa; T1) and 8 dpa (T2). These were selected as the time point when the first significant difference (P < 0.01, asterisks) in grain length was observed between 5A- (grey, dashed line, short grains) and 5A+ (purple, solid line, long grains) and the preceding time point. Differentially expressed (DE) transcripts were identified for four comparisons (q-value < 0.05). Coloured boxes indicate the numbers of DE transcripts identified for each comparison using alignments to either the IWGSC Chinese Spring Survey Sequence (CSS) or the TGACv1 (TGAC) Chinese Spring reference transcriptomes. Two ‘across time’ comparisons: (grey box; comparing T1 and T2 samples of the 5A- NIL) and (purple box; comparing T1 and T2 samples of the 5A+ NIL), and two ‘between NIL’ comparisons: (orange box; comparing 5A- and 5A+ NILs at T1) and (green box; comparing 5A- and 5A+ NILs at T2)
