Fig. 2.
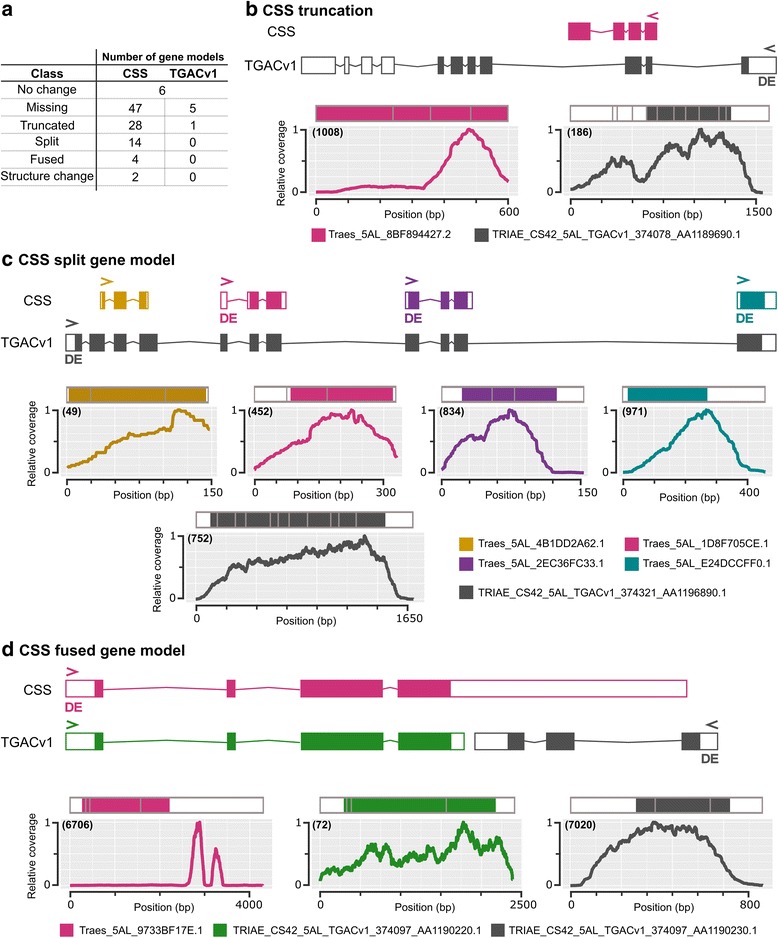
Comparison between CSS and TGACv1 gene models. a Discrepancies identified between gene models in the CSS and TGAC reference sequences and the number of gene models falling into categories. Panels (b), (c) and (d) show specific examples of discrepancies. In each panel, a representation of the unspliced gene model is shown with exons as coloured boxes, untranslated regions as white boxes, and introns as thin lines. Graphs show the relative read coverage across the spliced transcript with the structure represented diagrammatically directly above each graph. The number in brackets shows the maximum absolute read depth for each gene model. > and < in the gene structures indicate the direction of transcription and a ‘DE’ indicates that the gene model was differentially expressed in (q value < 0.05). For each panel transcript names are shown in the coloured legends
